Abstract
Cephenemyia stimulator parasitizes roe deer (Capreolus capreolus) throughout its geographical distribution. The complete circular C. stimulator mitogenome was assembled, which is 16,407 bp in length, and encodes 13 protein-coding genes, 22 tRNA genes, and 2 rRNA genes. A phylogenetic tree was built with mitogenome sequences, including C. stimulator and 13 related Oestridae species, using Sarcophaga tuberosa as an outgroup.
Genus Cephenemyia includes several species parasitizing cervid hosts. Cephenemyia stimulator (Hunter, 1916) can be found in the nasal cavity, pharynx, and throat of roe deer, Capreolus capreolus, throughout its geographical range (Zumpt Citation1965; Colwell et al. Citation2006; Scholl et al. Citation2019). In this study, we assembled and analyze for the first time the complete mitochondrial genome of C. stimulator.
Larvae of C. stimulator were obtained during the necropsy of a roe deer (Capreolus capreolus) (Lugo, Spain: 43°02′04.7′′N 7°38′31.1′′W). The specimens were deposited at the Department of Experimental Biology of the University of Jaén (https://www.ujaen.es/departamentos/bioexp; Contact email: [email protected]) under the references 713-1 to 713-10. Larvae were identified based on morphological criteria, in particular, the posterior peritremes and dorsal and ventral spinulation (Zumpt Citation1965; Colwell et al. Citation2006). The total DNA was extracted from half larvae of the specimen 713-5 with the Quick-DNA Tissue/Insect kit (Zymo Research) and 20 Gbp of sequences were obtained using the Illumina® Hiseq™ 2000 platform in paired-end reads with length 2 × 100 nt. We first performed a quality trimming with Trimmomatic (Bolger et al. Citation2014) to get complete read pairs with quality higher than Q20 in the whole read after removing adapters and used a million read pairs selected randomly with seqTK (https://github.com/lh3/seqtk). The mitogenome was assembled using the MITObim v1.8 program (Hahn et al. Citation2013) with the ‘–quick’ option with a maximum mismatch of 15%, and using as reference the mitogenome of Cephenemyia trompe (accession number MN814272; Li et al. Citation2020). The C. stimulator mitogenome was annotated with MITOS (Bernt et al. Citation2013) and tRNA scan-SE (Lowe and Eddy Citation1997), and annotations of the genes were refined by manual comparison with the C. trompe mitogenome (Li et al. Citation2020).
The length of the mitochondrial genome is 16,407 bp, with an overall 77.4% AT content (GenBank accession number MW145178). The nucleotide distribution for the mitochondrial genome is 39.5% A, 14.2% C, 8.4% G, and 37.9% T. It contains typically 37 genes (13 protein-coding genes, 22 transfer RNAs, 2 ribosomal RNAs) and one control region (D-loop). The mitochondrial genome of C. trompe is the only one available for the Cephenemyia genus (Li et al. Citation2020). In comparison, these two Cephenemyia mitogenomes are very similar in organizations and in length, 16,407 bp and 16,387 bp respectively for C. stimulator and C. trompe, and have a percentage of identity of 97.7%.
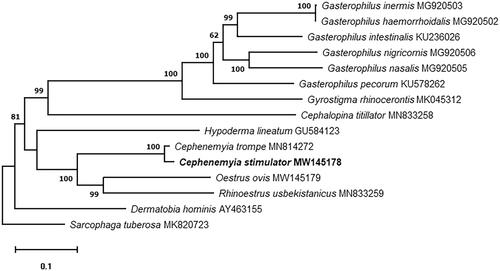
In addition, we analyzed the phylogenetic relationships of C. stimulator. The phylogenetic analysis was performed aligning the sequences with Clustal Omega (Sievers et al. Citation2011), the poorly aligned positions and divergent regions were removed using Gblocks program v0.91b (Talavera and Castresana Citation2007). The phylogenetic tree was built using the Maximum Likelihood method with 1000 replicates with MEGAX software (Kumar et al. Citation2018). For this analysis, the mitochondrial genome of the 13 Oestridae species currently available in GenBank were used, and the genome of Sarcophaga tuberosa (Zhang et al. Citation2019; MK820723) as outgroup. The results showed a conventional taxon pattern, previously established for the Oestridea family, and shows C. stimulator in the same branch that C. trompe, the sister group of the genera Estrus and Rhinoestrus ().
Acknowledgments
We would like to thank Dr. Francisco J. Ruiz-Ruano for the bioinformatics support.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The data that support the findings of this study are openly available in the US National Center for Biotechnology Information (NCBI database) at https://www.ncbi.nlm.nih.gov/, reference number: MW145178. Raw sequencing reads used in this study were deposited in the public repository BioSample with accession number SAMN19601264 (https://www.ncbi.nlm.nih.gov/biosample/19601264).
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120.
- Colwell DD, Hall MJR, Scholl PJ. 2006. A synopsis of the biology, hosts, distribution, disease significance and management. In: Colwell DD, Hall MJR, Scholl PJ, editors. The oestrid flies. Biology, host–parasite relationships, impact and management. Wallingford (UK): CABI Publishing; p. 220–305.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads—a baiting and iterative mapping approach. Nucleic Acids Research. 41(13):e129. doi:https://doi.org/10.1093/nar/gkt371.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGAX: Molecular Evolutionary Genetics Analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Li XY, Yan LP, Pape T, Gao YY, Zhang D. 2020. Evolutionary insights into bot flies (Insecta: Diptera: Oestridae) from comparative analysis of the mitochondrial genomes. Int J Biol Macromol. 149:371–380.
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25(5):955–964.
- Scholl PJ, Colwell DD, Cepeda-Palacios R. 2019. Myiasis (Muscoidea, Oestroidea). In: Mullen GR, Durden LA, editors. Medical and veterinary entomology, 3rd ed. San Diego (CA): Elsevier-Academic Press; p. 383–419.
- Sievers F, Wilm A, Dineen DG, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, et al. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 7(1):539.
- Talavera G, Castresana J. 2007. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 56(4):564–577.
- Zhang C, Shiwen W, Shang Y, Shen X, Guo Y. 2019. The complete mitochondrial genome of Sarcophaga brevicornis (Diptera: Sarcophagidae). Mitochondrial DNA B Resour. 4(2):2762–2763.
- Zumpt F. 1965. Myasis in man and animals in the old world. London (UK): Butterworths.