Abstract
In this study, we sequenced complete mitogenome of Taeniothrips tigris Bhatti 1995. It was 15,501 bp in length containing 13 protein-coding genes, 22 transfer RNA genes, and 2 ribosomal RNA genes along with two non-coding regions. The overall base composition of Ta. tigris is 43.66% A, 35.20% T, 11.46% C, and 9.68% G, with a high AT bias of 78.86%. The constructed phylogeny using 19 mitogenomes revealed that the genus Taeniothrips is in close relationship with genus Thrips. This mitogenome data would help in deducing phylogenetic relationships studies in the order Thysanoptera.
Keywords:
The members of the order Thysanoptera (Thrips) are minute, soft-bodied, and can be distinguished from the other insects by the presence of fringed wings and asymmetrical mouthparts (Mound and Marullo Citation1996). Out of 6200 known species, only 1% of thrips are reported as a serious pest (Thripswiki 2021). They are the sole transmitter of plant Tospoviruses, causing severe economic damage to a wide number of agricultural and horticultural crops (Riley et al. Citation2011; Tyagi et al. Citation2017). The genus Taeniothrips was first described by Amyot and Serville (Citation1843) with Type species Thrips primulae Haliday. Later on Bhatti (Citation1995) considered the Taeniothrips tigris as a type species of the genus Taeniothrips (Fetothrips). Ta. tigris is an endemic species to India and belongs to the most diverse subfamily Thripinae of the family Thripidae. This is the first report of mitogenome data for genus Taeniothrips in the order Thysanoptera.
The specimens of Ta. tigris were collected by a bush beating method in March 2020 at Nainital (29.38N, 79.45E), Uttarakhand, India. DNA was extracted by a nondestructive method (Tyagi et al. Citation2017) with DNeasy Blood and Tissue Kit (QIAGEN, Hilden, Germany -). All voucher specimens (registration No. 11077/H17-11104/H17) were deposited in the National Zoological Collections (NZC) at the Center for DNA Taxonomy, Molecular Systematics Division, Zoological Survey of India, Kolkata. The sequence data of Ta. tigris was generated on the Illumina platform NovaSeq 6000 (Illumina, San Diego, CA), with 2 × 150 base pair’s chemistry. Assembly was done by GetOrganelle software version 1.7.4 (Jin et al. Citation2020) and annotated in MITOS Web Server (Bernt et al. Citation2013), ORF Finder (https://www.ncbi.nlm.nih.gov/orffinder), BLASTn, and BLASTp and further compared with the thrips mitogenomes available on GenBank.
Ta. tigris mitogenome was 15,501 bp in length with 37 genes, including 13 protein-coding genes (PCGs), 22 transfer RNAs (tRNAs), 2 ribosomal RNAs (rRNAs), and 2 putative control regions (). Most of the genes were located on the majority strand except nad5, nad4, nad4L, trnH, trnP, and trnY. The AT content of the genome was 78.86% (43.66% of A, 35.20% of T) and GC 21.14% (9.68% of G, 11.46% of C). ATA start codon was used by cox2, cox3, cytb, nad2, nad4, nad5, atp8; ATT by cox1, nad1, nad3, nad6, atp6; and ATG by nad4L. All the PCGs were stopped with TAA stop codon with few exceptions. TAG stop codon was used by atp6, cox1; and incomplete termination codon T(AA) was used by nad2, nad3, nad4, and atp8. All the tRNAs had typical cloverleaf secondary structure with the length ranging from 71 bp (trnI) to 51 bp (trnV). The length of rrnL and rrnS were 1222 and 757 bp, respectively. Ta. tigris mitogenome contained 7 overlapping regions (1–7 bp with a total of 24 bp) and 21 intergenic spacer regions (1–165 bp with a total of 841 bp).
Figure 1. Phylogenetic tree inferred from 13 PCGs using Bayesian Inference. The posterior probabilities (pp) and bootstrap support (bs) are marked beside the nodes. The GenBank accession numbers are also provided. The hemipteran, Alloeorhynchus bakeri was used as an outgroup.
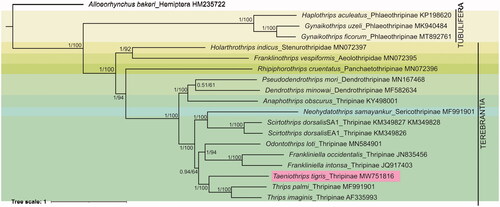
The PCGs were aligned with the MAFFT algorithm in TranslatorX tool (Abascal et al. Citation2010) and concatenated using SequenceMatrix version 1.8 (Vaidya et al. Citation2011). The best fit model was detected as ‘GTR + I + G’ in PartitionFinder version 2.1.1 using BIC criterion (Lanfear et al. Citation2017). The Bayesian inference (BI) and maximum likelihood (ML) phylogenetic trees were constructed using Mr. Bayes version 3.2 (Ronquist et al. Citation2012) and IQ tree web server (http://iqtree.cibiv.univie.ac.at/), respectively, with Alloeorhynchus bakeri (Genbank accession HM235722) as an outgroup. Both the phylogenetic methods produced similar tree topology and superimposed as shown in . The phylogeny revealed Ta. tigris was closely related to the genus Thrips (Th. palmi and Ta. imaginis) in subfamily Thripinae. The close relationship of the genus Taeniothrips and Thrips is also evidenced by the morphological characters as they do not have ocellar setae I. In contrast, the ctenidium on lateral side of tergites is present in Thrips and absent in Taeniothrips (Bhatti Citation1980; Mound et al. Citation2012). The complete mitogenome data of Ta. tigris will further broaden the knowledge gap in understanding the evolution, phylogeny, and gene arrangement in order Thysanoptera.
Acknowledgments
The authors are thankful to the Director of Zoological Survey of India (ZSI), Ministry of Environment, Forests and Climate Change (MoEFCC), Govt. of India for providing necessary permissions and facilities. This work is part of Ph.D. thesis of Avas Pakrashi.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference number MW751816.
Additional information
Funding
References
- Abascal F, Zardoya R, Telford MJ. 2010. TranslatorX: multiple alignment of nucleotide sequences guided by amino acid translations. Nucleic Acids Res. 38:W7–13.
- Amyot CJB, Audinet-Serville JG. 1843. Histoire Naturelle des Insectes. Hémiptères. Paris, Librairie encyclopédique de Roret. 1–657.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Bhatti JS. 1980. Species of the genus Thrips from India (Thysanoptera). System Entomol. 5(2):109–166.
- Bhatti JS. 1995. Further studies on Taeniothrips sensu lato (Insecta: Terebrantia: Thripidae). Zoology (Journal of Pure and Applied Zoology) 5:73–95.
- Jin JJ, Yu WB, Yang JB, Song Y, Depamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):31.
- Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B. 2017. Partitionfinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol Biol Evol. 34(3):772–773.
- Mound LA, Azidah AA, Ng YF. 2012. Key to the non-fossil species of the genus Taeniothrips (Thysanoptera, Thripidae). Zootaxa. 3414(1):33–42.
- Mound LA, Marullo R. 1996. The Thrips of Central and South America: an introduction. Mem Entomol Int. 6:1–488.
- Riley DG, Joseph SV, Srinivasan R, Dife S. 2011. Thrips vectors of tospoviruses. J Int Pest Manage. 1:1–10.
- Ronquist F, Teslenko M, Mark PV, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. Mrbayes 3.2: efficient bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542.
- Thrips Wiki. 2021. ThripsWiki – providing information on the World’s thrips. [accessed April 15, 2021]. https://thrips.info/wiki/.
- Tyagi K, Kumar V, Singha D, Chandra K, Laskar BA, Kundu S, Chakraborty R, Chatterjee S. 2017. DNA barcoding studies on Thrips in India: cryptic species and species complexes. Sci Rep. 7:1–14.
- Vaidya G, Lohman DJ, Meier R. 2011. SequenceMatrix: concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics. 27(2):171–180.
