Abstract
Liobagrus hyeongsanensis, Korean indigenous catfish, was reported as a new species in 2015. The complete mitochondrial DNA sequence of L. hyeongsanensis was sequenced by next-generation sequencing (NGS) analysis. The mitochondrial genome was assembled with 16,529 bp in length and encoded 13 protein-coding genes (PCGs), 22 tRNAs, two rRNAs, and one control region (D-loop). Also, the gene structures such as gene order and content were totally identical with the congeneric species. Molecular phylogenetic analysis determined the taxonomical position of L. hyeongsanensis in species level among the genus Liobagrus.
Genus Liobagrus inhabiting in South Korea is composed for five species, L. andersoni, L. obesus, L. mediadiposalis, L. somjinensis, and L. hyeongsanensis (Park and Kim Citation2010; Kim et al. Citation2015). Among these species, the complete mitochondrial genomes were analyzed except L. hyeongsanensis Kim et al. Citation2015 (Kartavtsev et al. Citation2007; Lee et al. Citation2016; Park et al. Citation2017; Kim et al. Citation2020). We analyzed the complete mitochondrial sequence of L. hyeongsanensis in this study and determined the molecular phylogenetic relationship between all of Liobagrus species from South Korea. Also, we examined the genome structure and characteristics, gene order and nucleotide base composition, and these results contributed to identify molecular features of the Liobagrus species.
Liobagrus hyeongsanensis specimen (voucher code: NIE-FI00001) used in mitochondrial genome analysis was collected from Buk River, Hwangnyong-dong, Gyeongju-si (35°51′10.03″N, 129°13′00.00″E) on 10 January 2021. A voucher specimen deposited at National Institute of Ecology (Seocheon, Korea), and mitochondrial DNA (mt-DNA) was stored in freezer (manager: Philjae Kim, [email protected], Seochen, Korea). Mitochondria was isolated from caudal fin using the Qproteome® Mitochondria Isolation Kit (QIAGEN, Hilden, Germany), and mt-DNA (NIE-DN00001) was extracted from mitochondria sample using DNeasy Blood and Tissue DNA isolation kit (QIAGEN). The extracted mt-DNA was stored in National Institute of Ecology until used. For NGS analysis, we obtained the PCR product from mt-DNA by using REPLI-g Mitochondrial DNA kit (QIAGEN). The library sample was prepared for sequencing analysis using QIAseq FX single cell DNA library kit (QIAGEN). The sequencing analysis was conducted by Illumina Hi-Seq 2500 platform (San Diego, CA, USA) in GnC Bio Co. (Daejeon, South Korea). The treatments of raw data, such as trimming, assembly and gene arrangement, were performed by using Geneious Prime 2021.1 (Biomatters, Auckland, New Zealand). The tRNA genes annotation and secondary structures were examined using tRNAscan-SE 2.0 (Chan and Lowe Citation2019).
The mitochondrial genome of L. hyeongsanensis (MZ066608) was 16,529 bp in length and composed of 13 PCGs, 22 tRNAs, two rRNAs, and one control region (D-loop). The gene order was completely identical as other Liobagrus species (Kartavtsev et al. Citation2007; Lee et al. Citation2016; Park et al. Citation2017; Kim et al. Citation2020). The nucleotide base composition was examined as 30.1% A, 25.3% T, 28.5% C, and 16.0% G. As with congeneric species, COI has initiation codon with ‘GTG’, and 12 of remainder PCGs started with ‘ATG.’ The six PCGs (ND2, COX2, COX3, ND3, ND4 and CytB) were used incomplete terminal codon ‘T––.’ The five PCGs (COX1, ATP8, ATP6, ND4L and ND5) and two PCGs (ND1 and ND6) have ‘TAA’ and ‘TAG’ as terminal codon, respectively.
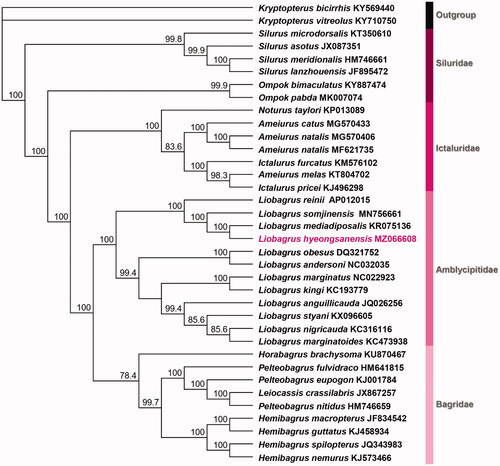
For determining the molecular phylogenetic relationship among the Korean Liobagrus species, we obtained the mitochondrial complete sequences of genus Liobagrus deposited in GenBank of NCBI (http://www.ncbi.nlm.nih.gov). All of the sequences were aligned with Geneious Prime 2021.1 (https://www.geneious.com). The dataset for phylogenetic analysis composed of 13 PCGs of a total of 36 Siluriformes mitochondrial genome. The maximum likelihood (ML) analysis with 1000 replicates conducted using PhyML 3.1, and best fit model was estimated by jModel Test with GTR + I + G substitution model (Guindon and Gascuel Citation2003; Guindon et al. Citation2010; Darriba et al. Citation2012). According to ML analysis, L. hyeongsanensis was placed on Liobagrus clade with congeneric species, and all of the family groups were formed monophyletic clade (). These result showed the phylogenetic status based on molecular and morphological evidences is stable in order Siluriformes. Our study would be a contributor for establishing the more precise phylogenetic relationship of Siluriformes.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MZ066608. The associated BioProject, SRA and Bio-Sample numbers are PRJNA717732, SRR14151707 and SAMN18509289 respectively.
Additional information
Funding
References
- Chan PP, Lowe TM. 2015. GtRNAdb 2.0: an expanded database of transfer RNA genes identified in complete and draft genomes. Nucleic Acids Res. 44:D184–D189.
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9(8):772.
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59(3):307–321.
- Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 52(5):696–704.
- Kartavtsev YP, Jung SO, Lee YM, Byeon HK, Lee JS. 2007. Complete mitochondrial genome of the bullhead torrent catfish, Liobagrus obesus (Siluriformes, Amblycipididae): Genome description and phylogenetic considerations inferred from the Cyt b and 16S rRNA genes. Gene. 396(1):13–27.
- Kim P, Han JH, An SL. 2020. Complete mitochondrial genome of Korean catfish, Liobagrus somjinensis (Actinopterygii, Siluriformes, Amblycipitidae), from South Korea. Mitochondrial DNA B Resour. 5(1):866–868.
- Kim SH, Kim HS, Park JY. 2015. A new species of torrent catfish, Liobagrus hyeongsanensis (Teleostei: Siluriformes: Amblycipitidae), from Korea. Zootaxa. 4007(2):267–275.
- Lee S, Kim JH, Song HY. 2016. Complete mitochondrial genome of the Korean torrent catfish Liobagrus andersoni (Siluriformes, amblycipitidae). Mitochondrial DNA B Resour. 1(1):779–780.
- Park CE, Kim MC, Kim KH, Park HC, Shin JH. 2017. The complete mitochondrial genome sequence of Liobagrus mediadiposalis (Teleostei, Siluriformes, Amblycipitidae). Mitochondrial DNA B Resour. 2(2):879–880.
- Park JY, Kim SH. 2010. Liobagrus somjinensis, a new species of torrent catfish (Siluriformes: Amblycipitidae) from Korea. Ichthyol Exploration Freshwaters. 21(4):345.