Abstract
The complete mitochondrial genome of Heteropriacanthus cruentatus has been obtained and annotated through Illumina next-generation sequencing. This mitogenome was found to be 16,506 bp in length, containing 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNA), and 2 ribosomal RNA genes (rRNA). This overall base composition of the complete mitogenome for this species included 27.52% A, 24.46% T, 16.99% G and 31.04% C. The phylogenetic analysis revealed that the H. cruentatus has the closest relationship with Pristigenys niphonia. This study provides an important resource for reviewing the phylogenetic relationships and taxonomic status of this species.
The family Pricanthidae is distributed in tropical and subtropical seas around the world near coral reefs or rock formations, which consists of 4 genera and 18 species of marine fishes known as bigeyes (Fernandez-Silva and Ho Citation2017). Heteropriacanthus cruentatus (Lacepède 1801) belongs to the family Pricanthidae and genus Heteropriacanthus, it has a wide distribution in Atlantic and Indo-Pacific oceans. This species is common in lagoons and seaward reefs with a usual depth between 3 and 35 m (Gasparini and Floeter Citation2001). Heteropriacanthus cruentatus was first used by Fitch and Crooke (Citation1984), and also has some synonymous name such as Priacanthus carolinus, P. cruentatus, and P. boops (Gaither et al. Citation2015). In this study, we reported the complete mitochondrial DNA sequence information of H. cruentatus and evaluated the relationship in comparison with closely related species.
The H. cruentatus samples were collected from the central coastal area of Zhejiang in East China Sea (27.0170°N, 123.0330°E). Sample of muscle tissue was collected from a specimen deposited at the East China Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences (Peng Sun, [email protected]) under a voucher number HCR2020801002. The total genomic DNA was extracted from a piece of muscle tissue using the DNA Extraction Kit (Tiangen, Beijing), and the extracted genomic DNA was amplified and sequenced through llumina NovaSeq platform using paired-end (2 × 150bp) sequencing mode by Shanghai Personal Biotechnology Co., Ltd. Then, the whole genome DNA was assembled by the SPAdes (Bankevich et al. Citation2012) and annotated by the MITOS (http://mitos.bioinf.uni-leipzig.de/) (Bernt et al. Citation2013).
The complete mitochondrial genome of H. cruentatus is 16,498 base pair (bp) in length (GenBank accession number: MZ042267) including 13 protein-coding genes, 2 ribosomal RNA (12S rRNA and 16S rRNA) and 22 transfer RNAs. Its base composition is 27.52% A, 24.46% T, 16.99% G and 31.04% C. Most of these protein-coding genes start translation with initiation codon of ATG, except for cox1 (ATC) and nd6 (ATT). The termination codons were TAA for genes of nd1, nd2, nd4, nd5, nd6 nd41, cox1, cox2, cox 3, atp6, atp8, cob, and TAG for genes of nd3. The rRNA position from 69 to 1016 with a length of 948 bp (rrnS), and from 1143 to 2789 with a length of 1647 bp (rrnL). The two non-coding regions are comprised of a light strand replication origin (OL) and D-loop region. The OL with a 33 bp in length is locate between trnN (gtt) and trnC (gac). The D-loop region (506 bp) is located between trnP (tgg) and trnF (gaa).
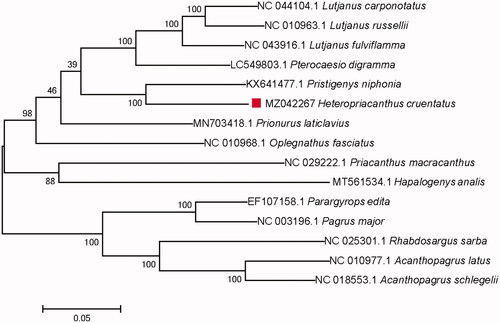
Fifteen mitochondrial genome sequences from Lutjanidae, Oplegnathidae, Sparidae, and Pricanthidae of Priacanthiformes and Eupercaria were used for phylogenetic construction by MEGA 7.0 with maximum likelihood method () and bootstrap value of 1000 replicates (Sudhir et al. Citation2016). Results showed that H. cruentatus has a closest phylogenetic relationship with Pristigenys niphonia. This study will provide useful genetic information to the genetic evolution and taxonomic study for H. cruentatus.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI (https://www.ncbi.nlm.nih.gov/) under the accession number MZ042267. The associated BioProject, SRA, and BioSample numbers are PRJNA724929, SRR14663579 and SAMN18865290, respectively.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Bernt M, Donath A, Juhling F, Externbrink F, Florentz C, Fritzsch G, Putz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Fernandez-Silva I, Ho HC. 2017. Revision of the circumtropical glasseye fish Heteropriacanthus cruentatus (Perciformes: Priacanthidae), with resurrection of two species. Zootaxa. 4273(3):341–361.
- Fitch JE, Crooke SJ. 1984. Revision of eastern Pacific catalufas (Pisces: Priacanthidae) with description of a new genus and discussion of the fossil record. Proc Calif Acad Sci. 43:301–315.
- Gaither MR, Bernal MA, Fernandez-Silva I, Mwale M, Jones SA, Rocha C, Rocha LA. 2015. Two deep evolutionary lineages in the circumtropical glasseye Heteropriacanthus cruentatus (Teleostei, Priacanthidae) with admixture in the south-western Indian Ocean. J Fish Biol. 87(3):715–727.
- Gasparini JL, Floeter SR. 2001. The shore fishes of Trindade Island, western South Atlantic. J Nat Hist. 35(11):1639–1656.
- Sudhir K, Glen S, Koichiro T. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.