Abstract
Camellia rostrata S. X. Yang & S. F. Chai is a recently described yellow camellia species from Guangxi, China. It is a critically endangered species according to the IUCN Red List Categories and Criteria. Here, we report the complete chloroplast (cp) genome based on next-generation sequencing technology. The complete cp genome of C. rostrata is 156,547 bp in length and consists of a large single-copy (LSC, 86,199 bp) region, a small single-copy (SSC, 18,204 bp) region, and a pair of inverted repeats (IRs, 26,072 bp). The genome contains 135 genes including 40 tRNA, eight rRNA, and 87 protein-coding genes. Phylogenetic analysis resolved C. rostrata in a clade containing C. huana and C. impressinervis, both of which are classified to Camellia sect. Archecamellia. Our findings support the placement of C. rostrata in C. sect. Archecamellia as proposed by a previous study. The cp genome of C. rostrata provides valuable bioinformatic resources for the protection and utilization of this yellow camellia species.
Camellia contains ca. 120 species (Ming Citation2000; Ming and Bartholomew Citation2007). It is mainly distributed in East and Southeast Asia, with a diversity centered in the Southern Yangtze River of China (Ming and Zhang Citation1996). One of these species, C. rostrata S. X. Yang & S. F. Chai is a newly described yellow species of Camellia from Guangxi, China (Liu et al. Citation2020). This species is restricted in its distribution to its type locality, which reportedly has <100 individuals, and was thus proposed as a critically endangered species according to the IUCN Red List Categories and Criteria (Liu et al. Citation2020). In this study, we report the complete cp genome sequence of C. rostrata, and constructed the phylogenetic relationship between C. rostrata and other congeneric species. The cp genome of C. rostrata provides useful bioinformatics for the conservation of this wild yellow species of Camellia.
Young leaves of C. rostrata were collected from Long’an county of Guangxi in China (23°03′58″N, 107°43′49″E). The voucher specimen (S. X. Yang & F. Y. Wu 6081) was deposited in the Herbarium at Kunming Institute of Botany (KUN), Chinese Academy of Sciences (KUN 1482804, http://www.kun.ac.cn, Jing-Hua Wang, [email protected]). Total genomic DNA was extracted using a modified hexadecyltrimethylammonium bromide (CTAB) approach (Doyle and Doyle Citation1987). Genome sequencing was performed using Illumina Hi-Seq 2500 platform. The chloroplast genome sequences were assembled using GetOrganelle (Jin et al. Citation2020) and annotated using PGA (Qu et al. Citation2019). Phylogenetic analysis of C. rostrata and 25 other species classified to Camellia as well as two outgroups was performed using RAxML version 8.2.12 (Stamatakis Citation2014) following a previous study (Yu et al. Citation2017).
The complete chloroplast genome of C. rostrata (GenBank accession number MW755303) was obtained with a total length of 156,547 bp and a mean sequencing depth of 155.2. The GC content of the genome is 37.3%. It consisted of a large single-copy (LSC, 86,199 bp) region, a small single-copy (SSC, 18,204 bp) region, and a pair of inverted repeats (IR, 26,072 bp). A total of 135 genes were annotated, including 87 protein-coding, 40 tRNA, and eight rRNA genes. Similar to cp genomes of other Camellia species from GenBank, it showed typical quadripartite structure reported in angiosperms (Shinozaki et al. Citation1986).
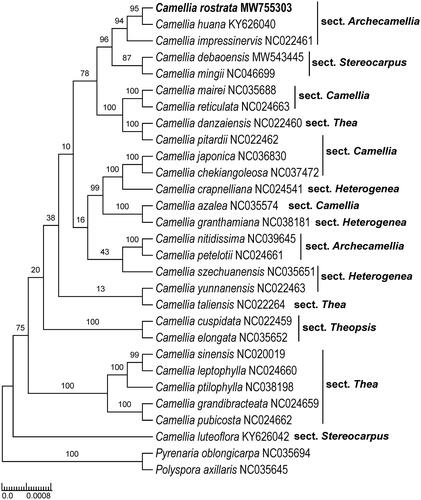
Based on morphological evidence, Liu et al (Citation2020) stated that C. rostrata was most closely related to C. debaoensis R. C. Hu & Y. Q. Liufu. However, these phylogenetic analyses indicate that C. rostrata, C. huana, and C. impressinervis grouped in a strongly supported clade (BS = 94%), which is sister to another highly supported clade containing C. debaoensis and C. mingii S. X. Yang (BS = 87%) (). Camellia huana and C. impressinervis are classified in C. sect. Archecamellia (Ming and Bartholomew Citation2007). Thus, our findings supported the placement of C. rostrata in C. sect. Archecamellia as proposed by Liu et al. (Citation2020). Both C. debaoensis and C. mingii were consistently placed in C. sect. Stereocarpus (Hu et al., Citation2019; Liu et al., Citation2019). Camellia sect. Archecamellia was not revealed as monophyletic since another one/two species (C. petelotii, C. nitidissima, the latter was treated as a synonym of the former according to Ming’s classification system) from this section were not nested within the C. rostrata clade. The cp genome reported here provides a reference for future study on the phylogenomics of Camellia, as well as the protection and utilization of C. rostrata.
Figure 1. Maximum likelihood tree of Theaceae based on 28 complete chloroplast genome sequences, including Camellia rostrata (GenBank accession number is MW755303) sequenced in this study. The bootstrap support values are shown beside the nodes. Two representative taxa of Theaceae (Polyspora axillaris, NC035645; Pyrenaria oblongicarpa, NC035694) were used as outgroups.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/, under the accession no. MW755303. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA725164, SRR14326646, and SRS8774895, respectively.
Additional information
Funding
References
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19(1):11–15.
- Hu RC, Wei SJ, Liufu YQ, Nong YK, Fang W. 2019. Camellia debaoensis (Theaceae), a new species of yellow camellia from limestone karsts in southwestern China. PhytoKeys. 135(135):49–58.
- Jin JJ, Yu WB, Yang JB, Song Y, dePamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241.
- Liu ZW, Chai SF, Wu FY, Ye PM, Jiang CJ, Zhang ZR, Yu XQ, Ma JL, Yang SX. 2020. Camellia rostrata, a new species of yellow camellias from Southwest China. Phytotaxa. 459(1):61–68.
- Liu ZW, Fang W, Liu ED, Zhao M, He YF, Yang SX. 2019. Camellia mingii, a new species of yellow camellias from Southeast Yunnan, China. Phytotaxa. 393(1):47–56.
- Ming TL. 2000. Monograph of the genus Camellia. Kunming: Yunnan Science and Technology Press.
- Ming TL, Bartholomew B. 2007. Theaceae. In: Wu CY, Raven PH, editors. Flora of China. Beijing; Saint Louis, MO: Science Press and Missouri Botanical Garden Press. p. 366–478.
- Ming TL, Zhang WJ. 1996. The evolution and distribution of genus Camellia. Acta Botanica Yunnanica. 18(1):1–13.
- Qu XJ, Moore MJ, Li DZ, Yi TS. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15: 50.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Shinozaki K, Ohme M, Tanaka M, Wakasugi T, Hayashida N, Matsubayashi T, Zaita N, Chunwongse J, Obokata J, Yamaguchi-Shinozaki K, et al. 1986. The complete nucleotide sequence of the tobacco chloroplast genome: its gene organization and expression. EMBO J. 5(9):2043–2049.
- Yu XQ, Gao LM, Soltis DE, Soltis PS, Yang JB, Fang L, Yang SX, Li DZ. 2017. Insights into the historical assembly of East Asian subtropical evergreen broadleaved forests revealed by the temporal history of the tea family. New Phytol. 215(3):1235–1248.
