Abstract
The first complete chloroplast genome of Aster ageratoides Turcz. var. scaberulus (Miq.) Ling. is reported in this study. The total chloroplast genome size of A. ageratoides var. scaberulus was 153,071 bp and comprised of a large single-copy region (LSC with 84,896 bp), a small single-copy region (SSC with 18,269 bp), and two inverted repeat regions (IR with 24,953 bp). A total of 122 genes were included in the genome, including 83 protein-coding genes, 8 rRNA genes, and 37 tRNA genes. Eleven protein-coding genes had intron (ycf3, clpP and rps12 gene contained two introns. Further phylogenomic analysis of Asteraceae, including 13 taxa, was conducted for the placement of A. ageratoides var. scaberulus.
Aster ageratoides Turcz. var. scaberulus (Miq.) Ling. (Asteraceae) is a perennial herb native to eastern Asia. It is widely distributed in China, Siberia Mongolia, South Korea and Japan (Chu et al. Citation2013). Because of its bright and daisy-like flowers, cultivars of A. ageratoides var. scaberulus are widely used as garden and cut flowers. Additionally, a recent study showed that aerial parts of A. ageratoides var. scaberulus are particularly rich in chlorogenic acid (CGA), which could potentially be used as a therapeutic agent in neurodegenerative disorders associated with glutamate excitotoxicity (Clifford et al. Citation2006; Rebai et al. Citation2017). Despite its importance in horticultural and medicinal value, there is little genetic information reported for A. ageratoides var. scaberulus. Here, we assembled and characterized the first complete chloroplast genome of A. ageratoides var. scaberulus. It will provide a valuable resource for further genetic conservation, evolution, and molecular breeding studies in the genus Aster.
Total DNA was extracted from dried leaves by DNA Plantzol Reagent (Invitrogen, Carlsbad, USA). The A. ageratoides var. scaberulus individual was collected from Chun’an, Zhejiang, China (GPS: E118°58′58.30″, N 29°46′56.71″). The specimen and extracted DNA were deposited at Zhejiang Province Key Laboratory of Plant Secondary Metabolism and Regulation, Zhejiang Sci-Tech University (http://sky.zstu.edu.cn) under the voucher number ZSTU00339 (collected by Zhe-Chen Qi, [email protected]). Whole plastome sequences were generated using the Illumina HiSeq-2500 platform (Illumina Inc., San Diego, CA, USA). In total, about 14.5 million high-quality clean reads (150 bp PE read length) were generated with adaptors trimmed. Aligning, assembly, and annotation was conducted by GetOrganelle (Jin et al. Citation2020) and GENEIOUS 11.1.5 (Biomatters Ltd, Auckland, New Zealand).
The full length of the A. ageratoides var. scaberulus chloroplast genome (GenBank Accession No. MW813970) was 153,071 bp and comprised of a large single-copy region (LSC with 84,896 bp), a small single-copy region (SSC with 18,269 bp), and two inverted repeat regions (IR with 24,953 bp). The overall GC content of the A. ageratoides var. scaberulus cp genome was 37.3% and the GC content of the LSC, SSC and IR regions are 35.2%, 31.3%, 43.1%. A total of 122 genes were included in the genome (83 protein-coding genes, eight rRNA genes, and 37 tRNA genes). Nineteen genes had two copies, which were comprised of seven PCG genes (ndhB, rpl2, rpl23, rps7, rps12, ycf1, ycf2), seven tRNA genes (trnV-GAC, trnR-ACG, trnN-GUU, trnL-CAA, trnI-GAU, trnI-CAU, trnA-UGC), and all five rRNA species (rrn16, rrn23, rrn4.5, rrn5, rrb16). In the genome, nine protein-coding genes (rps16, rpoC1, atpF, petB, petD, rpl16, rpl2, ndhB, ndhA) had one intron, and ycf3, clpP, rps12 gene contained two introns.
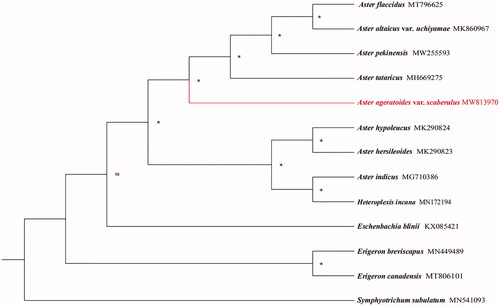
Twelve species with available chloroplast genomes were selected to study the phylogenetic placement of A. ageratoides var. scaberulus in Asteraceae. The alignment of plastomes was generated by MAFFT v7.475 (Katoh and Standley Citation2013). The maximum likelihood (ML) analysis was performed using IQTREE v 1.6.8 (Nguyen et al. Citation2015), of which the bootstrap values were calculated using 5000 replicates with the best-selected TVM + F+R2 model. The result showed that A. ageratoides var. scaberulus is sister to a clade formed by A. tataricus, A. pekinensis, A. ataicus and A. flaccidus according to the current sampling extent ().
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI (https://www.ncbi.nlm.nih.gov) under the accession no. MW813970. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA722057, SRR14241056, and SAMN18744330, respectively. The DNA matrix and phylogenetic tree that support the findings of this study are openly available in Github at https://github.com/andresqi/balsam-aster-plastome.
Additional information
Funding
References
- Chu SS, Liu LS, Liu ZQ, Jiang HG, Liu LZ. 2013. Chemical composition and insecticidal activities of the essential oil of Aster ageratoides flowering aerial parts. J Serb Chem Soc. 78(2):209–216.
- Clifford MN, Zheng W, Kuhnert N. 2006. Profiling the chlorogenic acids of aster by HPLC-MS(n). Phytochem Anal. 17(6):384–393.
- Jin J-J, Yu W-B, Yang J-B, Song Y, dePamphilis CW, Yi T-S, Li D-Z. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Rebai O, Belkhir M, Sanchez-Gomez MV, Matute C, Fattouch S, Amri M. 2017. Differential molecular targets for neuroprotective effect of chlorogenic acid and its related compounds against glutamate induced excitotoxicity and oxidative stress in rat cortical neurons. Neurochem Res. 42(12):3559–3572.