Abstract
Ricania shantungensis Chou & Lu, Citation1977 (Hemiptera: Ricaniidae), is an invasive pest that attacks forest as well as agricultural trees. We sequenced the 15,358 bp long complete mitochondrial genome (mitogenome) of this species; it consists of a typical set of genes (13 protein-coding genes, 2 rRNA genes, and 22 tRNA genes) and one major non-coding AT-rich region. The orientation and gene order of the R. shantungensis mitogenome are identical to that of the ancestral type found in majority of the insects. Bayesian inference (BI) phylogeny placed the R. shantungensis examined in our study, together with Ricania spp. in a group with the highest nodal support, forming the family Ricaniidae to which R. shantungensis belongs.
Ricania shantungensis Chou & Lu, Citation1977 (Hemiptera: Ricaniidae), is native to China (Chou and Lu Citation1977) and was detected as an exotic species in South Korea in 2011 (Rahman et al. 2011). Until now, host plants of R. shantungensis has been are known to be 138 species belonging to 62 families, including chestnut, peach, persimmon, apple, black locust, Japanese angelica tree, snowbell, pussy willow, paper mulberry, silk tree, tree of heaven, and Japanese cornlian cherry in Korea (Kim et al. Citation2015).
In previous studies, mitochondrial cytochrome c oxidase subunit I (COI) were analyzed from specimens collected from Korea; however, these specimens revealed that genetic difference was not associated with geographical distance, suggesting the need for variable markers for population genetics data (Kwon et al. Citation2017).
Materials and methods
One wild male adult sample was collected on the tree of heaven (Ailanthus altissima) by Hyobin Lee with an insect net in the Haenam-gun, Jeollanam-do, Republic of Korea (34.6724270 N, 126.617252 E) and its DNA was extracted from one of the hind legs. Leftover DNA and the specimen were deposited at the Gyeongsang National University, Jinju, Korea, under the accession number GNUCH08(M5). Using the extracted DNA, four long overlapping fragments (LF1: COI-ND3, LF2: ATP6-ND4, LF3: ND5-CytB, LF4: ND6-srRNA, LF5: IrRNA-COI) were amplified using four sets of primers designed using data regarding the previously published species of Ricaniidae (Ricania marginalis, NC_019597.1; Ricania speculum, NC_031369.1). Using the LFs as templates, 36 overlapping short fragments (SF) were amplified using the aforementioned primers.
The location and fragment size of protein coding genes (PCGs), tRNA genes, rRNA genes and AT-rich region were determined based on the published mitochondrial genomic of R. marginalis and R. speculum. tRNAscan-search server (http://lowelab.ucsc.edu/tRNAscan-SE/) and MITOS web server (http://mitos.bioinf.uni-leipzig.de/index.py) were applied to predict the tRNA genes secondary structure.
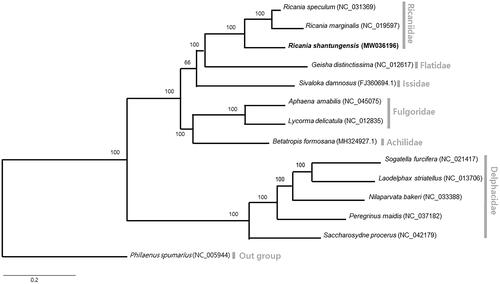
A phylogenetic analysis was performed using 13 available mitogenomes from Fulgoroidea, including the one obtained in this study (). Nucleotide sequences of all PCGs and rRNAs were aligned by using MAFFT (Kazutaka Citation2005), and 13 PCGs and 2 rRNAs were concatenated in alignment (15,023 bp). Bayesian inference (BI) method was applied using MrBayes version 3.2.6 (Ronquist et al. Citation2012). A phylogenetic tree was visualized using FigTree version 1.42 (http://tree.bio.ed.ac.uk/software/figtree/).
Results
The R. shantungensis mitogenome was found to be 15,358 bp in length, with typical gene sets − 2 rRNAs, 22 tRNAs, and 13 PCGs – and a major non-coding AT-rich region of 961 bp length (GenBank accession number MW036196). The gene arrangement of R. shantungensis was identical to that of the ancestral type found in majority of the insects (Boore Citation1999).
Phylogenetic analyses using a BI method, using 13 PCGs and 2 rRNAs, placed R. shantungensis from Korea, along with Ricania spp., into the genus Ricania, with the highest nodal support. The family Ricaniidae, to which R. shantungensis belongs, forms a cohesive monophyletic group with the highest nodal supports indicated by BI analysis.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MW036196. The associated BioProject and Bio-Sample numbers are PRJNA700097 and SAMN17823576, respectively.
Additional information
Funding
References
- Boore JL. 1999. Animal mitochondrial genomes. Nucleic Acids Res. 27(8):1767–1780.
- Chou I, Lu C. 1977. On the Chinese Ricaniidae with descriptions of eight new species. Acta Entomologica Sinica. 20(3):314–322.
- Kazutaka K. 2005. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 33(2):511–518.
- Kim DE, Lee H, Kim MJ, Lee D-H, Department of Eco-safety, Bureau of Conservation Ecology, National Institute of Ecology, Seocheon 325-813, Korea. 2015. Predicting the potential habitat, host plants, and geographical distribution of Pochazia shantungensis (Hemiptera: Ricaniidae) in Korea. Korean J Appl Entomol. 54(3):179–189.
- Kwon DH, Kim S-J, Kang T-J, Lee JH, Kim DH. 2017. Analysis of the molecular phylogenetics and genetic structure of an invasive alien species, Ricania shantungensis, in. Korea J. Asia Pac. Entomol. 20(3):901–906.
- Rahman M, Kwon Y, Suh S, Youn Y, Jo S. 2012. The genus Pochazia Amyot and Serville (Hemiptera: Ricaniidae) from Korea, with a newly recorded species. J of Entomology. 9(5):239–247.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542.