Abstract
The plant Cardamine amaraeformis Nakai. is considered an endemic plant to Korea. However, due to the similar morphological characteristics of C. amaraeformis with C. scutata, it is not easy to distinguish these two species. Here, the complete chloroplast genome of C. amaraeformis was sequenced and characterized. The chloroplast genome of C. amaraeformis was 155,598 bp in length, comprising a large single-copy (LSC) region of 84,574 bp and a small single-copy (SSC) region of 17,976 bp and a pair of inverted repeats (IRs) of 26,524 bp. The genome contained 131 genes, including 86 protein-coding (PCGs), 37 tRNA and 8 rRNA genes. Of those, 6 PCGs, 8 tRNA and 4 rRNA genes were duplicated in the IR region and one gene was a pseudogene. The GC content of the C. amaraeformis chloroplast genome was 36.3%. Phylogenetic analysis revealed that all Cardamine species formed a monophyletic clade and C. amaraeformis was closely associated with C. parviflora. Therefore, the present study could help to distinguish C. scutata and resolve the phylogenetic relationships among the Cardamine lineage.
The genus Cardamine belongs to the family of Brassicaceae that comprises more than 200 species (Al-Shehbaz Citation1988; Lihova and Marhold Citation2006). Specifically, in the Korean peninsula, sixteen species of Cardamine have been identified. Among these, C. amaraeformis Nakai, 1912. is considered an endemic plant to Korea (Park Citation2007). The morphological characteristics of the plant, C. amaraeformis contain cauline leaves with two pairs of leaflets; leaflets oblong or oval, nearly identical in size and shape, margins undulate or sinuate; petals obovate, white or pink, distinctly netted veined, 7–12 mm long (Ali et al. Citation2012). Moreover, the morphological characteristics of the species C. amaraeformis are very similar to the species C. scutata (Ali et al. Citation2012). Several molecular phylogenetic studies have been carried out to understand the phylogenetic relationship of the genus Cardamine using internal transcribed spacer sequences (Lihova and Marhold Citation2006; Ali et al. Citation2012). However, recent studies suggest that the plant chloroplast genome consists of highly conserved gene structures useful for resolving taxonomic and phylogenetic relationships (Al-Shehbaz Citation1988; Lihova and Marhold Citation2006; Ali et al. Citation2012). Hence, in the present study, we characterized the chloroplast genome of C. amaraeformis that may contribute to understanding the phylogenetic relationship of Cardamine in the Brassicaceae clade.
The fresh plant leaf materials were collected from mountain Cheongok, Bonghwa-gun, South Korea (geospatial coordinates: N37°4′9″, E128°57′47″) and the voucher specimen (YNUH21C062) was deposited at the Yeungnam University Plant Herbarium (http://lifesciences.yu.ac.kr), Gyeongsan, South Korea (Prof. SeonJoo Park, [email protected]). Total genomic DNA was extracted using a modified cetyltrimethylammonium bromide (CTAB) method (Doyle Citation1990) and deposited at the DNA Bank, Department of Life Sciences, Yeungnam University (http://www.yu.ac.kr/index.php), Gyeongsan, South Korea (Prof. SeonJoo Park, [email protected]). Whole-genome sequencing was achieved with a pair-end library (150 × 2) and an insert size of 350 base pairs (bp) using Illumina HiSeq 2500 sequencing system at LabGenomics, Seongnam, South Korea. Read quality was evaluated with FastQC v0.11.9 (Andrews Citation2010), and low-quality reads (LQR, <30%) were trimmed with Trimmomatic 0.40 (Bolger et al. Citation2014). The subsequent clean reads were filtered using the GetOrganelle v1.7.4.1 pipeline (https://github.com/Kinggerm/GetOrganelle) to acquire plastid-like reads and later, the filtered reads were assembled de novo method using SPAdes v3.15.2 (Prjibelski et al. Citation2020). Gene annotation was performed using Geneious Prime v2021.1.1. A circular chloroplast genome map was generated with OrganellarGenome DRAW (OGDRAW) (Greiner et al. Citation2019). The complete chloroplast genome sequence of C. amaraeformis and its gene annotation was submitted to GenBank (MZ043776).
The size of the C. amaraeformis Nakai, 1912. chloroplast genome was 155,598 bp with a typical quadripartite structure, including a large single-copy region (LSC, 84,574 bp) and a small single-copy region (SSC, 17,976 bp), separated by a pair of inverted repeats (IRs, 26,524 bp). This genome encoded 131 genes, including 86 protein-coding (PCGs), 37 tRNA and 8 rRNA genes. Out of 131 genes, 6 protein-coding, 8 tRNA and 4 rRNA genes were duplicated in the IR region and one gene was a pseudogene. The overall GC content of the chloroplast genome was 36.3%, while those of LSC, SSC and IR regions were 34.1, 29.2 and 42.4%, respectively. Moreover, the genome size, gene content and order were similar to other Cardamine chloroplast genomes.
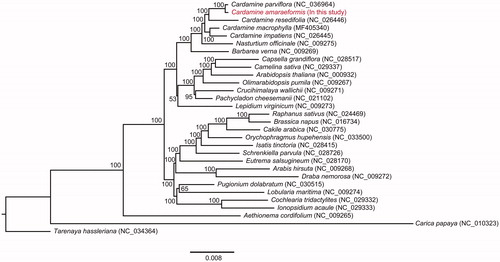
To understand the phylogenetic position of C. amaraeformis, the phylogenetic tree was constructed using 66 PCGs from 28 Brassicaceae and two outgroup chloroplast genomes. A maximum likelihood (ML) method, GTRGAMMA model and 1000 bootstrap replication were applied in the RAxML v8.2.X (Stamatakis Citation2014). Phylogenetic tree analysis revealed that all Cardamine species formed a monophyletic clade, and C. amaraeformis was closely related to C. parviflora (). Therefore, the C. amaraeformis chloroplast genome could be used to distinguish C. scutata and resolve the phylogenetic relationships within the Cardamine lineage.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in NCBI (https://www.ncbi.nlm.nih.gov) GenBank with the accession number MZ043776. The associated BioProject, SRA and Bio-Sample numbers are PRJNA738536, SRR14849702 and SAMN19732518, respectively.
Additional information
Funding
References
- Al-Shehbaz IA. 1988. The genera of Arabideae (Cruciferae, Brassicaceae) in the Southeastern United States. J Arnold Arbor. 69:85–166.
- Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data.
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120.
- Lihova J, Marhold K. 2006. Phylogenetic and diversity patterns in Cardamine (Brassicaceae) – a genus with conspicuous polyploid and reticulate evolution. In: Sharma AK and Sharma A, editors. Plant Genome: biodiversity and evolution. Enfield: Science Publishers; p. 149–186.
- Park C. 2007. The genera of vascular plants of Korea. Seoul: Academy Publishing Co.
- Ali MA, Lee J, Kim SY, Al-Hemaid FMA. 2012. Molecular phylogenetic study of Cardamine amaraeformis Nakai using nuclear and chloroplast DNA A markers. Genet Mol Res. 11(3):3086–3090.
- Doyle J. 1990. Isolation of plant DNA from fresh tissue. Focus. 12:13–15.
- Prjibelski A, Antipov D, Meleshko D, Lapidus A, Korobeynikov A. 2020. Using SPAdes de novo assembler. Curr Protoc Bioinformatics. 70(1):e102.
- Greiner S, Lehwark P, Bock R. 2019. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47(W1):W59–W64.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.