Abstract
Gammarus lacustris is native to the Qinghai–Tibet Plateau (QTP), widely distributed in alpine lakes. The complete mitochondrial DNA sequence of G. lacustris was 15,349 base pairs in length and comprised 13 protein-coding genes, 22 transfer RNA genes, 2 ribosomal RNA genes, and 1 control region. The BI tree showed that G. lacustris was most closely related to Gammarus duebeni, and indicated that Gammarus, Gmelinoides, Brachyuropus, Pallaseopsis, and Eulimnogammarus evolved from a common ancestor. The mitogenome of G. lacustris provides new molecular data for further taxonomic and phylogenetic studies of Amphipoda.
Gammarus lacustris (Amphipoda: Gammaridae) is a freshwater species, which is native to the Qinghai-Tibet Plateau (QTP) and widely distributed in alpine lakes (Hou and Li Citation2018). Gammarus lacustris has been found in Tibetan areas at altitudes of more than 5000 m, highly adapted to the extreme environment, such as strong UV radiation and hypoxia (Clewing et al. Citation2016). Gammarus lacustris is the food source for fishes and birds and maintains parasite diversity, playing an important role in the lacustrine food web (Shaw et al. Citation2020). The complete mitochondrial genome of G. lacustris provides an important resource for phylogenetic relationships and studying molecular evolution.
The G. lacustris sample was collected from Cuomujiri Lake (N 30.77°, E 90.79°), Tibet, Chian, and was stored in National Engineering Research Center for Marine Aquaculture, Zhejiang Ocean University (Jian, Chen, and [email protected]) under the voucher number GL20200610. Total genomic DNA was extracted from muscle using the DNeasy tissue kit (Qiagen). The mitochondrial sequences, amplified by PCR with 17 pairs of primer (Table S1), were obtained through Sanger dideoxy sequencing and assembled by CodonCode Aligner 5.1.5. The assembled mitochondrial genome was annotated using the online tool MITOS (Bernt et al. Citation2013). The annotated sequence was deposited in GenBank with the accession number MZ029704.
Similar to the mitogenomes of Gammarus, the complete mitochondrial genome of G. lacustris was 15,349 bp in length, including 13 protein-coding genes (PCGs), 22 tRNA genes, 2 rRNA genes (16S and 12S), and a control region (CR) (Cormier et al. Citation2018). The overall contents of A, T, C, and G were 31.83%, 32.55%, 22.58%, and 13.04%, respectively, with a high AT bias (64.38%). Both AT-skew and GC-skew of the mitogenome were negative, −0.0111 and −0.2643, respectively. The proportion of coding sequences with a total length of 11,066 bp was 72.10%, and 3679 amino acids were encoded. Except for four PSGs (ND1, ND4, ND4L, and ND5) and ten tRNA genes (Tyr, Gln, Cys, Phe, His, Pro, Leu1, Leu2, Val, and Ser), other mitochondrial genes were encoded on the H-strand. The lengths of 16S ribosomal RNA and 12S ribosomal RNA were 980 bp and 750 bp, respectively, which were both located in the positions between tRNA-Leu and CR, being separated by tRNA-Val. The length of CR was 984 bp, ranging from 14,366 bp to 15,349 bp.
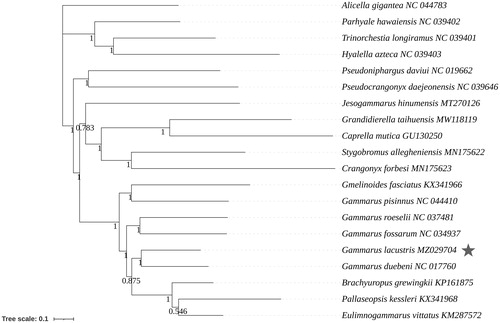
The phylogenetic relationships of G. lacustris within Amphipoda were reconstructed based on the 13 PSGs using the Bayesian inference (BI) phylogenetic tree. The BI tree was constructed by the software MrBayes 3.2.6 (Ronquist and Huelsenbeck Citation2003), with GTR + F + I + G4 as the best-fit evolutionary model determined by ModelFinder (Kalyaanamoorthy et al. Citation2017). The BI tree indicated that G. lacustris shared a close relationship with Gammarus duebeni (Figure 1). In addition, Gammarus species together with Gmelinoides fasciatus, Brachyuropus grewingkii, Pallaseopsis kessleri, and Eulimnogammarus vittatus formed a clade, representing these species evolved from a common ancestor (Figure 1). The complete mitogenome of G. lacustris presents here provides valuable resources for investigating the phylogenetic relationships within Amphipoda.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/nuccore/MZ029704) under the accession number: MZ029704.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Clewing C, Wilke T, Ilge A, Albrecht C. 2016. Phylogenetic patterns of freshwater amphipods inhabiting the Tibetan Plateau. Crustac. 89(2):239–249.
- Cormier A, Wattier R, Teixeira M, Rigaud T, Cordaux R. 2018. The complete mitochondrial genome of Gammarus roeselii (Crustacea, Amphipoda): insights into mitogenome plasticity and evolution. Hydrobiologia. 825(1):197–210.
- Hou Z, Li S. 2018. Four new Gammarus species from Tibetan Plateau with a key to Tibetan freshwater gammarids (Crustacea, Amphipoda, Gammaridae). ZooKeys, (747), 1.
- Kalyaanamoorthy S, Minh BQ, Wong TK, Von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589.
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19(12):1572–1574.
- Shaw JC, Henriksen EH, Knudsen R, Kuhn JA, Kuris AM, Lafferty KD, Siwertsson A, Soldánová M, Amundsen PA. 2020. High parasite diversity in the amphipod Gammarus lacustris in a subarctic lake. Ecol Evol. 10(21):12385–12394.