Abstract
The figleaf gourd (Cucurbita ficifolia Bouché), is a member of the Cucurbitaceae. Figleaf gourd genotypes are exclusively used as a rootstock for cucumber owing to their high physiological compatibility with cucumber. In this study, the complete chloroplast (cp) genome of C. ficifolia was assembled. The cp genome of C. ficifolia was 157,631 bp in length, it consists of a pair of inverted repeats (IRa and IRb) regions (25,638 bp) separated by the large single-copy (LSC, 88,211 bp) and small single-copy (SSC, 18,144 bp) regions. The cp genome encodes 111 unique genes, including 80 protein-coding genes, 27 transfer RNA genes, and four ribosomal RNA genes. The overall GC content of C. ficifolia cp genome was 37.2%. The phylogenetic tree of Cucurbitaceae showed that C. ficifolia was clustered into genus Cucurbita and the bootstrap value is 100%.
The figleaf gourd (Cucurbita ficifolia Bouché), which is synonymous with black-seed gourd, is a member of the Cucurbitaceae. It most likely originated in Central Mexico, subsequently spreading to South America, with an ecological preference for highland areas (Arriaga et al. Citation2006; Kim et al. Citation2010). The tender immature fruits, mature seed and young leaves of figleaf gourd are edible. Figleaf gourd genotypes are exclusively used as a rootstock for cucumber in Far Eastern and Western countries owing to their high physiological compatibility with cucumber (Roxas Citation1994; Lee and Oda Citation2003). While the figleaf gourd is highly tolerant to low temperature and salinity, it is susceptible to biotic stresses, such as nematodes and a Fusarium spp. (Lee and Oda Citation2003). To date, no genome information are available despite the potential value of this plant as a genetic resource for squash breeding. In the present study, the whole chloroplast (cp) genome of C. ficifolia was sequenced, assembled and annotated. Understanding the chloroplast genome information of this species may provide valuable guidelines for squash breeding.
The fresh leaves of C. ficifolia were collected from wild field of Mile county, Yunnan province of China (24.13°N, E103.31°E). The voucher specimen (Cuf6) was deposited at Laboratory of College of Horticulture and Landscape, Yunnan Agriculcural University (Voucher specimen: YNAU003512). Total genomic DNA was isolated from fresh leaves using a DNeasy Plant Mini Kit (QIAGEN, Valencia, California, USA) according to the manufacturer’s instructions to construction chloroplast DNA libraries. The Illumina sequencing was conducted by Shanghai Genesky Biotechnologies Inc. (Shanghai, China). Resultant clean reads were assembled using GetOrganelle pipeline (Jin et al. Citation2018, https://github.com/Kinggerm/GetOrganelle). The genome was automatically annotated by using the CpGAVAS pipeline (Liu et al. Citation2012) and start/stop codons and intron/exon boundaries were adjusted in Geneious R11.0.2 (Kearse et al. Citation2012). All the contigs were checked against the reference genome of C. maxima (NC_036505) and Cucurbita pepo (NC_038229). The raw data of sequence and annotation results were submitted to NCBI, under the accession number MW801448 and SRA number SRR14055461.
The complete cp genome sequence was 157,631 bp in length. It was the typical quadripartite structure and contained two short inverted repeat (IRa and IRb) regions (25,638 bp) which were separated by a small single copy (SSC) region (18,144 bp) and a large single copy (LSC) region (88,211 bp). The cp genome encodes 111 unique genes, including 80 protein-coding genes (PCGs), 27 transfer RNA (tRNA) genes, and four ribosomal RNA (rRNA) genes. Eighteen gene species are partially or completely duplicated, including seven PCGs, (ndhB, rpl2, rpl23, rps12, rps7, ycf15, ycf2), seven tRNAs (trnA-TGC, trnI-CAT, trnI-GAT, trnL-CAA, trnN-GTT, trnR-ACG, trnV-GAC), all four rRNAs (4.5S, 5S, 16S, 23S rRNA). The GC content of the cp genome was 37.2%, while the corresponding values of LSC, SSC, and IR regions was 34.9%, 31.6%, and 43.0%, respectively.
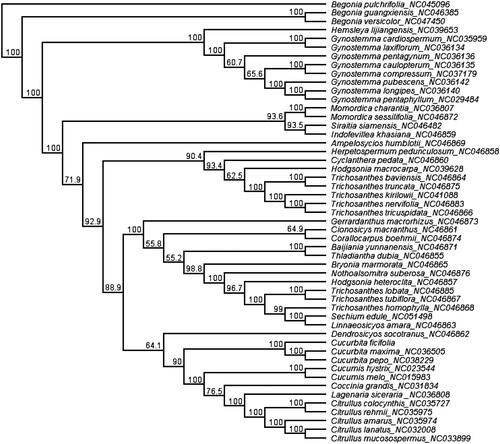
The 48 chloroplast genome sequences of Cucurbitaceae were downloaded from GenBank and aligned with C. ficifolia using MAFFT (Katoh and Standley Citation2013) in Geneious R11.0.2 (Kearse et al. Citation2012). To resolve its phylogenetic placement within the family Cucurbitaceae, the maximum likelihood (ML) phylogeny tree was reconstructed using IQtree (Lam-Tung et al. Citation2015). Begonia guangxiensis (NC_046385), Begonia pulchrifolia (NC_045096), and Begonia versicolor (NC_047450) from Begoniaceae were selected as outgroups. The topology of the phylogenetic tree showed that the species of C. ficifolia was clustered into genus Cucurbita (), the genus Cucurbita is a monophyletic group and the bootstrap value is 100%. The complete cp genome information reported in this study will be a valuable resource for future studies of the species’ genetic diversity, and breeding research of squash.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MW801448. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA716427, SRR14055461, and SAMN18435723, respectively.
Additional information
Funding
References
- Arriaga L, Huerta E, Lira-Saade R, Moreno E, Alarcón J. 2006. Assessing the risk of releasing transgenic Cucurbita spp. in Mexico. Agr Ecosyst Environ. 112(4):291–299.
- Jin JJ, Yu WB, Yang JB, Song Y, Yi TS, Li DZ. 2018. GetOrganelle: a simple and fast pipeline for de novo assembly of a complete circular chloroplast genome using genome skimming data. BioRxiv. 4:256479.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Kim KM, Kim CK, Han JS. 2010. In vitro regeneration from cotyledon explants in figleaf gourd (Cucurbita ficifolia Bouch), a rootstock for Cucurbitaceae. Plant Biotechnol Rep. 4(2):101–107.
- Lam-Tung N, Schmidt HA, Arndt VH, Quang MB. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Lee JM, Oda M. 2003. Grafting of herbaceous vegetable and ornamental crops. Hort Rev. 28:61–124.
- Liu C, Shi L, Zhu Y, Chen H, Zhang J, Lin X, Guan X. 2012. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genomics. 13(1):715.
- Roxas VP. 1994. Cucurbita ficifolia Bouché. In: Siemonsma JS, Piluek K, editors. Plant resources of South-East Asia 8 vegetables, Bogor Indonesia. Wageningen: Pudoc; p. 165–167.