Abstract
Pseudo-nitzschia is an important genus of diatoms with many species capable of inducing harmful algae blooms (HABs) in coastal and oceanic waters, some of which produce the toxin domoic acid (DA), a neurotoxin that causes amnesic shellfish poisoning (ASP). Pseudo-nitzschia delicatissima is a cosmopolitan species that can induce HABs and produce DA. Nevertheless, mitochondrial genome of P. delicatissima has not been revealed. In this study, we determined the complete mitochondrial genome of P. delicatissima for the first time. The circular mitochondrial genome was 42,182 bp in length with GC content of 30.37%. It consisted of 65 genes including 39 protein-coding genes (PCGs), 24 tRNA genes, and two rRNA genes. This mitogenome has a group II intron, located in the cytochrome c oxidase subunit genes (cox1), with orf790 identified inside the intron region. Phylogenetic analysis revealed that P. delicatissima was clustered well with P. multiseries. This analysis is valuable for studying the evolutionary relationships among Pseudo-nitzschia species, and for comparative analysis of P. delicatissima strains.
Diatoms are a widespread and diverse group of unicellular eukaryotic algae, and they have significant ecological importance in the carbon and silicate cycles, accounting for approximately 20% of the global photosynthetic carbon fixation (Field et al. Citation1998). Pseudo-nitzschia is a species-rich genus of diatoms, many of which can induce harmful algae blooms (HABs) in coastal and oceanic waters and produce the toxin domoic acid (DA), a neurotoxin causing amnesic shellfish poisoning (ASP) (Bates et al. Citation2018). The species Pseudo-nitzschia delicatissima (Cleve) Heiden 1928 has been identified to exist in many regions including Europe, America, Asia, and Oceania (Bates et al. Citation2018). Furthermore, P. delicatissima has been reported to bloom in Gulf of Naples (Italy), Berowra Creek (Australian), Bay of Seine (France), Bizerte Lagoon (Tunisia), Jeddah (Saudi Arabia), Fujian Province (China), South China Sea, and some strains can produce DA (Orsini et al. Citation2004; Li Citation2012; Fernandes et al. Citation2014; Thorel et al. Citation2017; Al-Aidaroos et al. Citation2019; Ajani et al. Citation2020). At present, morphological features of P. delicatissima has been characterized. It has lanceolate-shaped cells with 1.5–2.0 μm in width and 19–76 μm in length, containing two plastids and being capable of forming stepped chains (Lundholm et al. Citation2006; Ghiron et al. Citation2008). Nevertheless, mitochondrial genome of P. delicatissima has not been revealed. In this study, we determined and analyzed the complete mitochondrial genome of P. delicatissima.
The strain CNS00135 was isolated from water samples collected from the Jiaozhou Bay, China (36°02'058″N, 120°22'866″E) in October 2019 onboard the research vessel “Innovation”. It was identified to be P. delicatissima based on its morphological features and sequence similarity of known molecular markers including rbcL, 18S rDNA, and ITS. Previous studies had found large genetic variation among the strains of P. delicatissima, and many new species were separated from P. delicatissima group (Amato et al. Citation2007; Quijano-Scheggia et al. Citation2009). In this study, ML tree based on ITS showed that CNS00135 belong to the same clade with strain Laesø5, which was the epitype of P. delicatissima (Lundholm et al. Citation2006), thus CNS00135 was identified to be P. delicatissima sensu stricto. The voucher samples were deposited at the KLMEES of IOCAS (Nansheng Chen, [email protected]) under the voucher number CNS00135.
Total DNA was extracted using the modified CTAB method (Doyle Citation1987). DNA sample were sequenced using the Illumina NovaSeq 6000 platform (Illumina, San Diego, CA, USA). The mitochondrial genome was de novo assembled using the program GetOrganelle (Jin et al. Citation2020), with SPAdes version 3.10.1 as assembler (Bankevich et al. Citation2012). Subsequently, the obtained genome sequence was checked by aligning sequencing reads against the mitochondrial genome using the MEM algorithm of BWA v0.7.17 (Li and Durbin Citation2010). Alignments were visualized using IGV v2.8.12 (Robinson et al. Citation2011). ORF finder (https://www.ncbi.nlm.nih.gov/orffinder) and MFannot (https://megasun.bch.umontreal.ca/RNAweasel/) were used to annotate the mitochondrial genome of P. delicatissima.
The complete circular mitochondrial genome (mtDNA, GenBank accession No: MW436413) of P. delicatissima was 42,182 bp in length, which was shorter than that of P. multiseries (46,283 bp), the only mitochondrial genome of a Pseudo-nitzchia species that has been published (Yuan et al. Citation2016). The GC content of P. delicatissima was 30.37% (T 33.52%, C 15.59%, A 36.11%, and G 14.78%), which was lower than that of P. multiseries (31.05%). The mtDNA of P. delicatissima consisted of 65 genes including 39 PCGs (including three open reading frames, orfs), 24 tRNA genes, and two rRNA genes. Among the 39 PCGs, the start codons 35 genes were ATG, while the start codons of atp8, cob, rpl5, and orf790 were TTG, TTG, ATT, ATC, respectively. The stop codons of 32 PGGs were the canonical stop codon TAA, while the stop codons of cox1, nad4L, nad6, nad9, tatC were TAG, respectively, and the stop codons of rpl5 and orf790 were AAA and GGA, respectively. A group II intron was identified in the mtDNA of P. delicatissima, located in the cytochrome c oxidase subunit genes (cox1). A putative gene orf790 was found within the intron in the cox1 gene. Comparative analysis of the mtDNAs of P. delicatissima and P. multiseries revealed that P. multiseries has 3 group II introns in the cox1 gene (NC_027265) based on our re-annotation, with 3 ORFs (orf714, orf742, orf790) inserted in these introns of the cox1 gene.
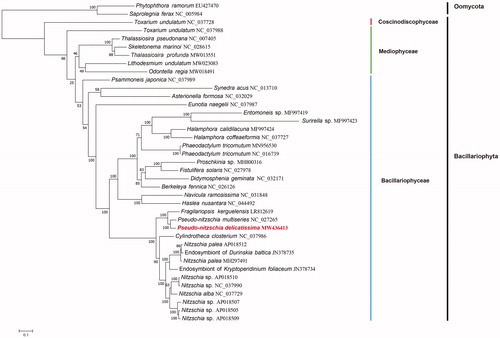
A Maximum Likelihood (ML) phylogenetic tree () was constructed using IQtree v1.6.12 (Jana et al. Citation2016) with 1000 bootstrap alignments based on tandem amino acid sequences of 31 common PCGs (including atp6, 8, 9; cob; cox1, 2, 3; nad1-7, 4L, 9, 11; rpl2, 5, 6, 14, 16; rps3, 4, 8, 10, 11, 13, 14, 19; and tatC) obtained from 36 publicly diatom mitochondrial genomes, including Odontella regia (MW018491), Lithodesmium undulatum (MW013551), and Thalassiosira profunda (MW013551) that we have recently constructed (Liu et al. Citation2021; Wang et al. Citation2021). Phytophthora Ramorum (EU427470) and Saprolegnia Ferax (NC_005984) were used as out-group taxa in the phylogenetic tree. As shown in , all diatoms species were divided nicely into three clades corresponding to three classes of Coscinodiscophyceae, Mediophyceae, and Bacillariophyceae. As expected, P. delicatissima clustered well with P. multiseries with high support (100% bootstrap). Additionally, P. delicatissima and P. multiseries were clustered with Fragilariopsis kerguelensis, indicating a closer evolutionary relationship between Pseudo-nitzschia and Fragilariopsis. In conclusion, the complete mitochondrial genome of P. delicatissima could provide important information for understanding the phylogeny and evolution of diatoms.
Acknowledgements
We are thankful to all members of Marine Ecological and Environmental Genomics Research Group at the Institute of Oceanology, the Chinese Academy of Sciences. We are grateful to colleagues at the Jiaozhou Bay Marine Ecosystem Research Station for their assistance with sampling during the expedition.
Disclosure statement
The authors are responsible for the content and writing of the paper. The authors report no conflicts of interest.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MW436413. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA693874, SRR13500717, and SAMN17487522, respectively.
Additional information
Funding
References
- Ajani PA, Larsson ME, Woodcock S, Rubio A, Farrell H, Brett S, Murray SA. 2020. Fifteen years of Pseudo-nitzschia in an Australian estuary, including the first potentially toxic P. delicatissima bloom in the southern hemisphere. Estuarine Coastal Shelf Sci. 236:106651.
- Al-Aidaroos AM, Devassy RP, El-Sherbiny MM. 2019. Unusual dominance of harmful microalgae Pseudo-nitzschia delicatissima cf.(Cleve) Heiden in the coastal waters of Jeddah, central Red Sea. Pak. J. Bot. 51(2):1–6.
- Amato A, Kooistra WH, Ghiron JHL, Mann DG, Pröschold T, Montresor M. 2007. Reproductive isolation among sympatric cryptic species in marine diatoms. Protist. 158(2):193–207.
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Bates SS, Hubbard KA, Lundholm N, Montresor M, Leaw CP. 2018. Pseudo-nitzschia, Nitzschia, and domoic acid: new research since 2011. Harmful Algae. 79:3–43.
- Doyle J. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Fernandes LF, Hubbard KA, Richlen ML, Smith J, Bates SS, Ehrman J, Léger C, Mafra LL, Kulis D, Quilliam M, et al. 2014. Diversity and toxicity of the diatom Pseudo-nitzschia Peragallo in the Gulf of Maine, Northwestern Atlantic Ocean. Deep Sea Res 2 Top Stud Oceanogr. 103:139–162.
- Field CB, Behrenfeld MJ, Randerson JT, Falkowski P. 1998. Primary production of the biosphere: integrating terrestrial and oceanic components. Science. 281(5374):237–240.
- Ghiron JHL, Amato A, Montresor M, Kooistra WH. 2008. Plastid inheritance in the planktonic raphid pennate diatom Pseudo-nitzschia delicatissima (Bacillariophyceae). Protist. 159(1):91–98.
- Jana T, Lam-Tung N, Arndt v. H, Quang MB. 2016. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. W1:W232–W235.
- Jin J-J, Yu W-B, Yang J-B, Song Y, Depamphilis CW, Yi T-S, Li D-Z. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):231–241.
- Li H, Durbin R. 2010. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 26(5):589–595.
- Li X-D. 2012. Analysis on characteristics of red tide in Fujian coastal waters during the last 10 years. Huan Jing Ke Xue. 33(7):2210–2216.
- Liu K, Liu S, Chen Y, Liu F, Chen N. 2021. Complete mitochondrial genome of the harmful algal bloom species Thalassiosira nordenskioeldii (Mediophyceae, Bacillariophyta) from the East China Sea. Mitochondrial DNA Part B. 6(4):1421–1423.
- Lundholm N, Moestrup Ø, Kotaki Y, Hoef‐Emden K, Scholin C, Miller P. 2006. Inter-and intraspecific variation of the Pseudo-nitzschia delicatissima complex (Bacillariophyceae) illustrated by rRNA probes, morphological data and phylogenetic analyses 1. J Phycol. 42(2):464–481.
- Orsini L, Procaccini G, Sarno D, Montresor M. 2004. Multiple rDNA ITS-types within the diatom Pseudo-nitzschia delicatissima (Bacillariophyceae) and their relative abundances across a spring bloom in the Gulf of Naples. Mar Ecol Prog Ser. 271:87–98.
- Quijano-Scheggia SI, Garcés E, Lundholm N, Moestrup Ø, Andree K, Camp J. 2009. Morphology, physiology, molecular phylogeny and sexual compatibility of the cryptic Pseudo-nitzschia delicatissima complex (Bacillariophyta), including the description of P. arenysensis sp. nov. Phycologia. 48(6):492–509.
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. 2011. Integrative genomics viewer. Nat Biotechnol. 29(1):24–26.
- Thorel M, Claquin P, Schapira M, Le Gendre R, Riou P, Goux D, Le Roy B, Raimbault V, Deton-Cabanillas A-F, Bazin P, et al. 2017. Nutrient ratios influence variability in Pseudo-nitzschia species diversity and particulate domoic acid production in the Bay of Seine (France). Harmful Algae. 68:192–205.
- Wang Y, Chen Y, Wang J, Liu F, Chen N. 2021. Mitochondrial genome of the harmful algal bloom species Odontella regia (Mediophyceae, Bacillariophyta). J Appl Phycol. 33(2):855–868.
- Yuan X-L, Cao M, Bi G-Q. 2016. The complete mitochondrial genome of Pseudo-nitzschia multiseries (Baciuariophyta). Mitochondrial DNA A DNA Mapp Seq Anal. 27(4):2777–2778.