Abstract
Paris polyphylla var. alba is a medicinal plant commonly used in the southwest of China. This study characterized the complete chloroplast (cp) genome sequence of P. polyphylla var. alba to investigate its phylogenetic relationship in Melanthiaceae. The cp genome of P. polyphylla var. alba is 165,079 base pairs (bp) in length with 36.96% G + C content. The cp genome is divided into (a) large single copy (LSC) (84,393 bp), (b) small single copy (SSC) (16,066 bp), and (c) two inverted regions (32,310 bp). The cp genome contains 135 genes, including 89 protein-coding genes, 38 tRNA genes, and 8 rRNA genes. Phylogenetic analysis indicated that P. polyphylla var. alba is closest to P. polyphylla var. emeiensis, and Paris had a close relationship with Trillium in Melanthiaceae.
Paris polyphylla var. alba H.Li & R.J.Mitchell is a flowering herb belonging to the Paris genus in the Melanthiaceae family, first described by Li Heng in 1986 (Li Citation1986; Wu et al. Citation2018). The dried rhizome has been widely used in traditional Chinese medicine for the treatment of hemostatic, sore throat, snake bite, and convulsion, etc. (Duan et al. Citation2018, Fu et al. Citation2012, Liu et al. Citation2017). To date, the studies of this species have mainly focused on its pharmacological activity, chemical composition, and quantitative analysis (Yin et al. Citation2007, Zhe et al. Citation2017). However, no data are available regarding the genomic studies on P. polyphylla var. alba and its relationship with other species belonging to Paris. Herein, the cp genome of Paris polyphylla var. alba was assembled and characterized for the first time, which will provide helpful information for further study of the genus.
Fresh leaves of P. polyphylla var. alba collected from Germplasm Resource Garden of Longmen Township (Yunan, China, 25°32′38″N, 99°32′8″E). A specimen was deposited at the herbarium of Dali University (https://www.dali.edu.cn/, Baozhong Duan and [email protected]) under the voucher number 20200820A11. A total genomic DNA of sample was extracted with plant genomic DNA kit (Tiangen Biotech, China) and sequenced by using the Hiseq 2500 platform (Illumina, San Diego, CA). Approximately 4.15 Gb of raw data (26,636,980 reads) was assembled by NOVOPlasty (Nicolas et al. Citation2017), and annotated by CPGAVAS2 (Shi et al. Citation2019). Annotated cp genome sequence was submitted to GenBank under accession number no. MW727455.
The cp genome sequence of P. polyphylla var. alba is 165,079 bp in length, with a large single-copy region (LSC) of 84,393 bp, a small single-copy region (SSC) of 16,066 bp, and a pair of inverted repeats (IR) regions of 32,310 bp. GC content of the whole genome, LSC, SSC, and IRs regions are 36.96%, 35.65%, 31.57%, and 40.00%, respectively. A total of 135 genes were annotated, including 89 protein-coding genes, 38 tRNA genes, and 8 rRNA genes. The gene content and organization of the inverted repeat are similar to the other members of Paris (Fan et al. Citation2020).
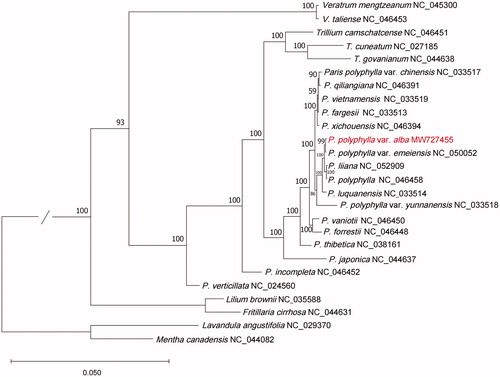
To reveal the phylogenetic position of P. polyphylla var. alba with other members of Melanthiaceae, a phylogenetic analysis was performed based on 23 complete cp genomes. In addition, Lavandula angustifolia (NC_029370) and Mentha Canadensis (NC_044082) were downloaded and included as an outgroup. The MAFFT v7.307 was used to extract the coding sequences, and a total of 81 coding sequences (accD, atpA, atpB, atpE, atpF, atpH, atpI, ccsA, cemA, clpP, infA, lhbA, matK, ndhA, ndhB, ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, ndhK, petA, petB, petD, petG, petL, petN, psaA, psaB, psaC, psaI, psaJ, psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbT, psbZ, rpcL, rpl2, rpl14, rpl16, rpl20, rpl22, rpl23, rpl32, rpl33, rpl36, rpoA, rpoB, rpoC1, rpoC2, rps2, rps3, rps4, rps7, rps8, rps11, rps12, rps14, rps15, rps16, rps18, rps19, ycf1, ycf2, ycf3, ycf4, ycf15) were presented in all of the 24 species. Then the MAFFT v7.307 was used to concatenate the coding sequences and align the concatenation sequences. Afterward, RAxML (version 8.2.12) (Stamatakis Citation2014) was used to construct the maximum likelihood (ML) tree; bootstrap probability values were calculated from 1000 replicates. The ML tree showed that P. polyphylla var. alba, P. polyphylla var. emeiensis, P. liiana, and P. polyphylla were clustered together (). The work reported the first complete cp genome of P. polyphylla var. alba, which will provide further insight into the evolution of Melanthiaceae.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The data that support the findings of this study are openly available in GenBank at https: https://www.ncbi.nlm.nih.gov/nuccore/MW727455, Associated BioProject, https://www.ncbi.nlm.nih.gov/bioproject/PRJNA728587, BioSample accession number at https://www.ncbi.nlm.nih.gov/biosample/SAMN19092376 and Sequence Read Archive at https://www.ncbi.nlm.nih.gov/sra/SRR14506499.
Additional information
Funding
References
- Duan BZ, Wang YP, Fang HL, Xiong C, Li XW, Wang P, Chen SL. 2018. Authenticity analyses of Rhizoma paridis using barcoding coupled with high resolution melting (Bar-HRM) analysis to control its quality for medicinal plant product. Chin Med. 13(13):8–10.
- Fan M, Liu J, Wang J, Qian J, Xia C, Duan B. 2020. The first complete chloroplast genome sequence of Paris polyphylla var. emeiensis, a rare and endangered species. Mitochondrial DNA B Resour. 5(3):2172–2173.
- Fu SZ, Li N, Liu Z, Gao WY, Man SL. 2012. Determination of seven kinds of steroidal saponins in plants of Paris L. from different habitats by HPLC. Chin Tradit Herb Drugs. 43(12):2435–2437.
- Li H. 1986. A study on the taxonomy of the genus Paris L. Bull Bot Res. 6:123–124.
- Liu Y, Luo D, Yao H, Zhang X, Duan B. 2017. A new species of Paris sect. Axiparis (Melanthiaceae) from Yunnan, China. Phytotaxa. 4(2):3278–3279.
- Nicolas D, Patrick M, Guillaume S. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 4(45):e18.
- Shi L, Chen H, Jiang M, Wang L, Wu X, Huang L, Liu C. 2019. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47(W1):W65–W73.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Wu XM, Zuo ZT, Zhang QZ, Wang YZ. 2018. Classification of Paris species according to botanical and geographical origins based on spectroscopic, chromatographic, conventional chemometric analysis and data fusion strategy. Microchem J. 143(2018):367–378.
- Yin HX, Zhang H, Xue D. 2007. Paris polyphylla var. emeiensis H. X. Yin, H. Zhang & D. Xue, a new variety of Trilliaceae from Sichuan. China. Acta Phytotaxon Sin. 45(06):822–827.
- Zhe WU, Wang YZ, Zhang J, Yang SB, Zhang JY, Fu-Rong XU. 2017. Study on genetic relationship of Paris polyphylla var. yunnanensis and its wild relatives based on infrared spectroscopy. Chin Tradit Herb Drugs. 48(11):2279–2284.