Abstract
Rhododendron mole (Blume) G. Don is an attractive ornamental and valuable medicinal plant which widely distributed in the southern regions of China. In order to promote the studies on the genetic diversity of this species, we assembled the complete chloroplast (cp) genome of R. molle by using the genome skimming approach. The results showed that the cp genome of R. molle exhibited a quadripartite cycle with 197,877 bp, comprising of two inverted repeats (IRs) of 43,831 bp separated by a large single copy (LSC) region of 110,189 bp and a quite small single copy (SSC) region of 26 bp. It encodes 146 genes, including 92 protein-coding, 46 tRNA, and eight rRNA genes. The overall GC content of the cp genome was 36.0%. The phylogenetic analysis indicated that R. molle is closely related to R. delavayi. Thus, the cp genome sequence of R. molle provides a rich source of genetic information for studies on Rhododendron taxonomy, phylogeny, and evolution, as well as lays the foundation for further development and utilization of R. molle.
Rhododendron molle (Blume) G. Don (Ericaceae) is a deciduous shrub, which is mainly distributed in the southern regions of China (Wu et al. Citation2005). With flourishing branches and gorgeous flowers, it is one of the elite parents for new varieties improvement in Rhododendron. Moreover, previous studies suggested that different organs and tissues of R. molle (including flower and root, etc.) could be used as analgesics and insecticides (Zhong et al. Citation2006; Li et al. Citation2020; Zhang et al. Citation2020). Due to lacking genome information and available molecular markers, the phylogenetic relationship of R. molle with other related species is still unclear. Herein, we assembled the cp genome of R. molle and inferred its phylogenetic position in family Ericaceae which will be conducive to promoting the studies on the phylogeography, genetic diversity, and utilization of this species.
Fresh leaves of R. molle were collected from Changsha, Hunan, China (28°6′3′′N 113°12′22′′E, 75 m). The voucher specimen (CSFI036611) was deposited at Central South University of Forestry and Technology (https://www.cvh.ac.cn/spms/detail.php?id=f0cda901, Lei Wu, [email protected]; [email protected]). The genomic DNA was isolated using the DNeasy Plant Mini Kit (QIAGEN GmbH, Hilden, Germany). A paired-end library of 2 × 150 bp and insert size of ∼350 bp was constructed according to the Illumina standard library, which was then sequenced on the Illumina HiSeq 2500 platform at Genepioneer Biotechnologies Inc. (Nanjing, China). About 4.9 GB clean data were used to assemble the cp genome using SPAdes (Bankevich et al. Citation2012). The cp genome of R. delavayi (MN711645.3) was used as a reference for further adjustments and then annotated using Dual Organellar Genome Annotator (DOGMA) (Wyman et al. Citation2004). The sequence of cp genome was deposited in GenBank (accession number: MZ073672).
The cp genome of R. molle was 197,877 bp in length, consisting of a pair of inverted repeats (IRs), a large single copy (LSC), and a small single copy (SSC), and the sequence lengths were 43,831 bp, 110,189 bp, and 26 bp, respectively. The overall GC content of the cp genome was 36.0%, while the GC percentages in IR, LSC, and SSC were 36.9, 35.3, and 7.7%, respectively. The genome encodes 146 genes, including 92 protein-coding, 46 tRNA, and eight rRNA genes.
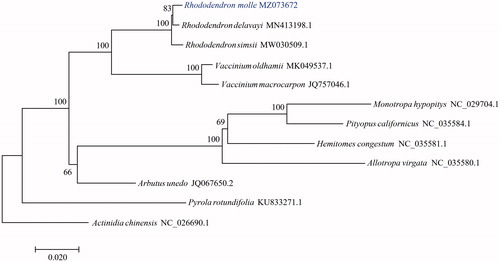
To investigate the phylogenetic position of R. molle, the cp genome of 11 species belong to Ericaceae and Actinidiaceae (outgroup) were obtained. Coding sequences shared by all species were multiple aligned with MAFFT (Katoh and Standley Citation2013) and connected. The maximum-likelihood (ML) tree was reconstructed using RAxML (Stamatakis Citation2014) with the GTRGAMMA model and 1000 bootstrap replicates. The result showed that R. molle was closely related to R. delavayi ().
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank, National Center for Biotechnology Information (NCBI) at https://www.ncbi.nlm.nih.gov/genbank/, with accession number of MZ073672. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA742763, SRR15010628, and SAMN19967795, respectively.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Li Y, Zhu YX, Zhang ZX, Li L, Liu YB, Qu J, Ma SG, Yu SS. 2020. Antinociceptive grayanane-derived diterpenoids from flowers of Rhododendron molle. Acta Pharm Sin B. 10(6):1073–1082.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Wu ZY, Peter R, Hong DY. 2005. Flora of China: Apiaceae through Ericaceae. Vol. 14. Beijing: Science Press; St. Louis: Missouri Botanical Garden Press; p. 242–517.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255.
- Zhang HP, Zhu YX, Zhang ZX, Chai LS, Liu YB, Yu HB, Li Y. 2020. New triterpenoids from the roots of Rhododendron molle as positive modulators of GABAA receptors. Tetrahedron. 76(37):131455.
- Zhong GH, Liu JX, Weng QF, Hu MY, Luo JJ. 2006. Laboratory and field evaluations of rhodojaponin-III against the imported cabbage worm Pieris rapae (L.) (Lepidoptera: Pieridae). Pest Manag Sci. 62(10):976–981.