Abstract
Trikeraia hookeri is an alpine grass with significant ecological value. Here, the complete chloroplast genome sequence of T. hookeri using Illumina sequencing data was reported. The size of the whole cp genome was 137,696 bp in length, consisting of a pair of inverted repeats (IR 13,755 bp), a large single-copy region (LSC 81,613 bp), and a small single-copy region (SSC 28,568 bp). The T. hookeri chloroplast genome encodes 119 genes: 81 mRNA genes, 34 tRNA genes and 4 rRNA genes. The GC content of T. hookeri chloroplast genome was 38.8% and those in LSC, SSC, and IR regions were 36.9, 40.8, and 42.3%, respectively. The maximum-likelihood phylogenetic analysis demonstrated that T. hookeri was most closely related to Stipa lipskyi (NC028444) and Stipa purpurrea (NC029390). Our findings provide fundamental information for further evolutionary and phylogenetic researches of T. hookeri.
Poaceae is an important vegetation consistute part in alpine grassland community. The Stipeae, belonging to the subfamily Pooideae, family Poaceae, which is of special significance among the alpine steppe vegetation in Qinghai-Tibet plateau, which include approximately 400–500 species and 24 genera in all of the world, in which 67 species and 10 genera were distributed in china. Trikeraia hookeri is a kind of Stipeae, extremely salinity-tolerant, cold-tolerant and drought-tolerant species (Khan Citation2003), distributed at 4300–5400 m in Qinghai and Tibet of china (Shukla and Srivastava Citation2020). T. hookeri increased the coverage of plant community, also provided food and habitation for animals. However, no studies on the plastome of T. hookeri have been published. In this study, we reported the complete chloroplast genome sequence of T. hookeri and explored its internal relationships with the family steppe.
The seeds of T. hookeri was collected from Zhongba county (83°30′05″E, 30°10′38″N), Shigatse, Tibet of China. A specimen was deposited at Northwest Institute of Plateau Biology, Chinese Academy of Sciences (NWIPB, Sha Xiao, [email protected]) under the voucher number 0334882. In 2020, the seed was grown in light incubator, at four-leaf stage, the leaf was extracted DNA by CTAB (Dreisigacker et al. Citation2012) and then sequenced using the Illumina NovaSeq platform (Illumina, San Diego, CA). The raw data were used to assemble the complete cp genome using GetOrganelle software (Jin et al. Citation2020), and the chloroplast genome of related species Nassella hyalina (GeneBank accession number NC036696) were used as the reference genome. The genome annotation was p erformed with the program Geneious R8 (Biomatters Ltd, Auckland, New Zealand). Finally, the complete chloroplast genome sequences of T. hookeri were submitted to GenBank (accession number: MW699773).
The whole chloroplast genome of T. hookeri cp genome is 137,696 bp in size, consisting of a pair of inverted repeats (IR 13,755 bp), a large single-copy region (LSC 81,613 bp), and a small single-copy region (SSC 28,568 bp). A total of 119 genes were annotated, containing 81 mRNA genes, 34 tRNA genes and 4 rRNA genes. The GC content of T. hookeri chloroplast genome was 38.8% and those in LSC, SSC, and IR regions were 36.9, 40.8, and 42.3%, respectively.
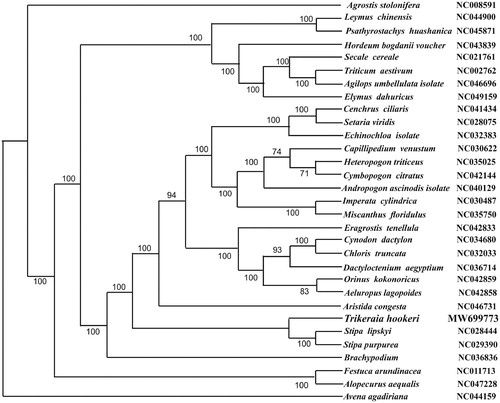
To confirm the phylogenetic location of T. hookeri, 11 genera of complete chloroplast genomes from Gramineae were obtained from GenBank, and the 12 complete chloroplast sequences were aligned using MAFFT (Katoh and Standley Citation2013). The maximum-likelihood analysis was performed by MEGA7 (Kumar et al. Citation2016) with bootstrap set to 1,000 (). The phylogenetic analysis showed a strong sister relationship with Stipa lipskyi (NC028444) and Stipa purpurrea (NC029390).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MW699773. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA671634, SAMN16539309, and SRR12888103, respectively.
Additional information
Funding
References
- Dreisigacker S, Sehgal D, Luna B, Reyes AE, Mollins J. 2012. CIMMYT wheat molecular genetics laboratory protocols and applications to wheat breeding. Mexico: CIMMYT.
- Jin JJ, Yu WB, Yang JB, Song Y, dePamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Khan MA. 2003. An ecological overview of halophytes from Pakistan. In: Kratochwil A, Lieth H, editors. Cash crop halophytes: recent studies. Netherlands: Kluwer Academic Publishers; p. 175.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Shukla N, Srivastava SK. 2020. Flora of Ladakh: an annotated inventory of flowering plants. In: Ghulam HD, Anzar AK, editors. Biodiversity of the Himalaya: Jammu and Kashmir State. Springer Nature Singapore Pte Ltd. Singapore: Springer Nature; p. P.724.