Abstract
We sequenced the complete chloroplast genome of Asplenium komarovii Akasawa (syn: Asplenium scolopendrium L. subsp. japonicum (Komarov) Rasbach, Reichstein & Viane), which is designated as a rare species in South Korea. The complete chloroplast genome is 149,393 bp in total length and comprised of the following regions: large single copy (82,464 bp), small single copy (21,345 bp), and a pair of inverted repeats (22,792 bp). The overall GC content is 40.9% and the genome encoded a total of 115 genes, including 84 protein-coding, 27 transfer RNA, and 4 ribosomal RNA genes. Phylogenetic analysis based on 21 representative chloroplast genomes of the suborder Aspleniineae (and one outgroup) indicates that Aspleniaceae is monophyletic and sister to Diplaziopsidaceae, with Rhadidosoraceae as the basal group in this three family clade. Asplenium komarovii is sister to A. nidus and A. prolongatum with strong bootstrap support. The chloroplast genome of A. komarovii will be useful in establishing its relationships within the A. scolopendrium complex, which is currently unresolved.
Asplenium komarovii Akasawa (synonym: Asplenium scolopendrium L. subsp. japonicum (Komarov) Rasbach, Reichstein & Viane) is distributed in Korea, Japan, China, and Russia. In Korea, it is found in the inland regions of Gangwon-do and Jeolla-do, and the two islands of Ulleung and Jeju (Ok and Yoo Citation2012), but most populations are very small and fragmentated. Therefore, A. komarovii has been protected as a rare plant by the Korea Forest Research Institute since 1997.
Asplenium scolopendrium is a temperate evergreen fern species, which occurs in Europe, North America, and East Asia. The taxonomic history of this species is extensive and contentious, and currently with 5 major classifications: two European (A. scolopendrium var. scolopendrium and A. scolopendrium subsp. antri-jovis), two American (A. scolopendrium var. americanum and A. scolopendrium var. lindenii), and one Asian (A. scolopendrium subsp. japonicum). The complexity of their variations with respect to morphology, ploidy levels (2x in var. scolopendrium; 4x in var. americanum and subsp. japonicum), ecological preferences, and geographic distribution has created considerable scientific interest as well as taxonomic ambiguity (Emmott Citation1964; Futyma Citation1980; Cinquemani et al. Citation1988). Even if morphological characters show differences among varieties or subspecies, they are usually not decisive. This indicates that successful taxonomic treatment of the A. scolopendrium complex requires multiple criteria. Apparently, even with the current used of both morphology and ploidy level screening, the ambiguity in identification and classification remains. Thus, the incorporation of additional traits is necessary and use of DNA markers will provide decisive identification and classification, and such can be achieved with the complete chloroplast genome sequence of members of the A. scolopendrium complex. This report on the chloroplast genome of A. komarovii is going to pave the way for the establishment of its phylogenetic relationships with other Asplenium species, particularly the members of the A. scolopendrium complex. Information on the chloroplast genome of A. komarovii is also necessary to provide new insights for future studies such as marker development for inter- and intra-species identification, population genetics, and reconstruction of evolutionary history.
Here, we report the complete chloroplast genome of A. komarovii. We obtained a leaf sample from the Korea National Arboretum (voucher specimen KHB1469365, N33°24′1″ E126°24′39″). DNA was isolated using the DNeasy Plant mini kit (Qiagen, Germany) and sequenced with the Illumina platform (Macrogen, Korea). A total of 71,538,770 reads were obtained and assembled de novo with Velvet v. 1.2.10 using multiple k-mers (Zerbino and Birney Citation2008). The genes for annotation were identified using DOGMA (Wyman et al. Citation2004 ) and tRNAscan-SE software with default settings (Schattner et al. Citation2005). The start and stop codons were manually corrected and further verified by homology searches using BLAST (Altschul et al. Citation1990). The complete chloroplast genome of A. komarovii has been deposited in GenBank under the accession number MZ064529. For phylogenetic analysis, the chloroplast genome of A. komarovii and 21 other species belonging to the Aspleniineae were used with Hypodematium crenatum from Polypodiineae as the outgroup. Maximum-likelihood analysis was conducted using IQ-TREE v.1.6.7 (Nguyen et al. Citation2015).
The chloroplast genome of A. komarovii is 149,393 bp in size. It exhibits a circular quadripartite structure comprised of large single copy (LSC) and small single copy (SSC) regions, which are 82,464 and 21,345 bp long, respectively. They are separated by a pair of inverted repeats (IRA and IRB) which are each 22,792 bp long. The GC contents of the LSC, SSC, and IR regions are 39.7, 37.7, and 44.7%, respectively. Asplenium komarovii has a total of 115 unique genes which are subdivided into 84 protein-coding, 27 tRNA and four rRNA genes.
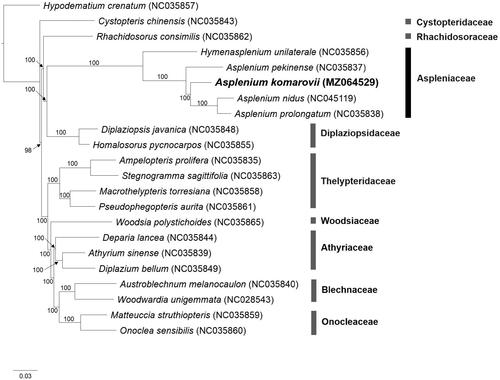
The phylogenetic tree shows that Aspleniaceae is monophyletic and sister to Diplaziopsidaceae, with Rhachidosoraceae as the basal group among these three families that form as a clade (), which is supported by strong bootstrap values (BS = 100). This clade is distinct from the rest of the Aspleniineae, which also have mostly strong bootstrap supports (BS = 100). These results are generally consistent with the previous studies (Cui et al. Citation2019; Lehtonen and Cárdenas Citation2019). Asplenium species shared a recent common ancestor (BS =100) and that A. komarovii is closely related to A. nidus and A. prolongatum (). The relationship of A. komarovii with members of the A. scolopendrium complex will be examined with the availability of the complete chloroplast genome sequence from A. scolopendrium var scolopendrium and var. americanum (Heo, Yun and Fernando, unpublished data).
Acknowledgement
We thank the Korea National Arboretum for providing the sample for Asplenium komarovii.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession no. MZ064529. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA726739, SRR14381415, and SAMN18957913, respectively.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol. 215(3):403–410.
- Cinquemani DM, Faust ME, Leopold DJ. 1988. Periodic censuses (1916–1986) of Phyllitis scolopendrium var. americana in central New York State. Am Fern J. 78(2):37–43.
- Cui C, Hong Y, Liu S, Wang Z, Wang T, Su Y. 2019. Complete chloroplast genome sequence of Asplenium nidus (Aspleniaceae), an economically important foliage fern. Mitochondr DNA Part B. 4(1):923–924.
- Emmott JI. 1964. A cytogenetic investigation in a Phyllitis‐Asplenium complex. New Phytol. 63(3):306–318.
- Futyma RP. 1980. The distribution and ecology of Phyllitis scolopendrium in Michigan. Am Fern J. 70(3):81–87.
- Lehtonen S, Cárdenas GG. 2019. Dynamism in plastome structure observed across the phylogenetic tree of ferns. Bot J Linn Soc. 190(3):229–241.
- Nguyen LT, Schmidt HA, Von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Ok GH, Yoo KO. 2012. Habitats ecological characteristics of Asplenium scolopendrium L. and its RAPD analysis. Korean J Plant Res. 25(6):719–730.
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33(Web Server issue):W686–W689.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18(5):821–829.