Abstract
Rhodobryum laxelimbatum (Bryophyta, Bryaceae) is one of the folk medicine resources in Southwest China, which has excellent potential for application in treating cardiovascular diseases. In this study, R. laxelimbatum was sequenced by high-throughput sequencing technology. The complete chloroplast genome is 124,632 bp in length with a quadripartite structure. Two inverted repeat regions are 9837 bp, separated by a large single copy region of 86,444 bp and a small single copy region of 18,514 bp. It encodes 118 unique genes, including 82 protein-coding genes, 32 tRNA genes, and four rRNA genes. The phylogenetic tree was constructed based on the complete chloroplast genome sequences of 18 bryophytes, downloaded from GenBank and acquired in this study. The phylogenetic analysis strongly indicated that R. laxelimbatum was the sister group of the clade which consists of Mnium marginatum, Pohlia cruda and Pohlia nutans. The R. laxelimbatum chloroplast genome sequence provides new genomic resources, which will improve its research, conservation, and application in the future.
The genus of Rhodobryum was established by German botanist Limpricht (Citation1892) and was placed in the family of Bryaceae. Rhodobryum got the name because its leaves were clustered at the stem’s tip in a distinct rose-shaped pattern (Rhodon means rose, and Bryum is the type genus of Bryaceae). The Chinese name of Rhodobryum, ‘Da-ye-xian’, means mosses with big leaves because the leaves of Rhodobryum are larger than other Bryaceae species’ (Chen Citation1963). As a traditional folk medicine, the species of the genus Rhodobryum are broadly popular known to treat cardiovascular disease and nervous prostration (Li Citation1990; Hu et al. Citation2009, Li and Zhao Citation2009; Cai et al. Citation2011). Several pharmacological studies have attested to their efficacy (Harris Citation2008; Harris and Yang Citation2009), yet little is known about the chloroplast genome sequences of Rhodobryum. Therefore this study of R. laxelimbatum will help for better understanding of this genus and provide valuable information for the phylogeny of Bryaceae (Shi et al. Citation2021).
DNA of Rhodobryum laxelimbatum was extracted from dry leaves by a modified CTAB method (Li et al. Citation2013). The sample was collected from Haba Snow Mountain in Shangri-La, Yunnan, China (100.117°E, 27.313°N), and the voucher specimen (Shuo Shi, SZ5212; September 19, 2016) was deposited in the herbarium of Hebei Normal University, HBNU. The people in charge of the HBNU collection is Shuo Shi, email: [email protected]. With a paired-end (PE 150) genomic library acquired by the Illumina HiSeq X Ten platform (by Novogene Biotech Company, Beijing), the R. laxelimbatum complete chloroplast genome was assembled with Spades (Bankevich et al. Citation2012) and Geneious (Kearse et al. Citation2012), the boundaries among the repetitive and unique regions, we confirm such regions by mapping original reads (150 bp long) to the corresponding region. The sequence was annotated in DOGMA (Wyman et al. Citation2004) and edited in Sequin v15.50.
The complete chloroplast DNA sequence of R. laxelimbatum (GenBank accession No. MW147233) is 124,632 bp in length. Two inverted repeat regions (IRs) are 9837 bp long and separated by a large single copy (LSC) region of 86,444 bp and a small single copy (SSC) region of 18,514 bp. The genome contains 118 unique genes, including 82 protein-coding genes, 32 tRNA genes, and four rRNA genes (Detail information is available at GenBank: https://www.ncbi.nlm.nih.gov/nuccore/MW147233.1/#feature_MW147233.1). Through gene annotation, 15 genes (trnL-GAU, trnA-UGC, ndhA, ndhB, rpl2, rpl16, petD, petB, trnV-UAC, trnL-UAA, trnK-UUU, trnG-GCC, atpF, rpoC1, ycf66) contain one intron separately, and three genes (clpP, ycf3 and rps12) each possess two introns. The overall G/C content is 29.22%. Moreover, there are 158 simple sequence repeats (SSR) in the chloroplast genome. SSRs were detected by MISA (MicroSatellite identification tool) with the search parameters set at the minimum repeat threshold of mononucleotide 10 bp, dinucleotide 12 bp, trinucleotide, tetranucleotide and pentanucleotide 15 bp, and hexanucleotide 24 bp.
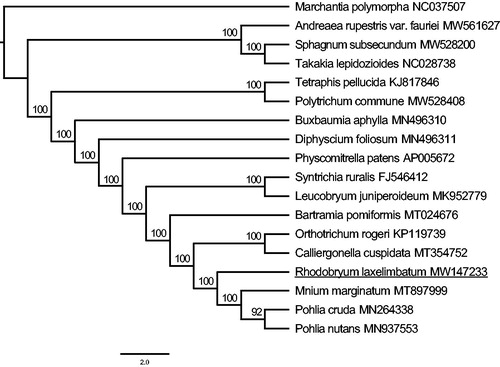
The ML tree was constructed with PhyloSuite v1.1.14 (Zhang et al. Citation2020) based on the complete chloroplast genomes of 17 mosses and one liverwort (Marchantia polymorpha, as outgroup). As we all know, there are majorly two types of chloroplast genome data deposited in the NCBI, contributing to the different orientations among the repetitive regions (IRa and IRb) and unique regions (LSC and SSC). Firstly, we used Mauve (Darling et al. Citation2004) to determine the gene collinearity of the chloroplast genome and manually adjust the specific structure of the genome. Then the whole genome data of 18 genomes were aligned by the MAFFT v7.222 (Katoh and Standley Citation2013) and manually adjusted with BioEdit v7.0.9.0 (Alzohairy Citation2011). Phylogenetic analysis was conducted based on maximum-likelihood analyses implemented in IQ-TREE (Nguyen et al. Citation2015) with a TVM + R3 + F model for 10,000 ultrafast (Minh et al. Citation2013) bootstraps, as well as the Shimodaira-Hasegawa-like approximate likelihood-ratio test (Guindon et al. Citation2010). The tree was edit in Figtree v1.4.3. In the tree (), most clades were supported with high bootstrap values (100%) and R. laxelimbatum was the sister group of the clade which consists of Mnium marginatum, Pohlia cruda and Pohlia nutans.
Geolocation information
The study area of this research is in Yunnan, China.
Acknowledgment
We thank Dr. Shuo Shi for his suggestions and technical support in this manuscript. We thank Dr. Wenpan Dong and Dr. Yanlei Liu for their assistance in data analyses. We thank Mr. Ibrahim M. Ahmad for helping to revise the manuscript.
Disclosure statement
The authors report no conflict of interest.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession number MW147233. The associated BioProject, SRA, and Bio-Sample number are PRJNA725408, SRR14332107 and SAMN18878621.
Additional information
Funding
References
- Alzohairy AM. 2011. Bioedit: an important software for molecular biology. GERF Bull Biosci. 2(1):60–61.
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, et al. 2012. Spades: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Cai Y, Lu Y, Chen RH, Wei QL, Lu XH. 2011. Anti-hypoxia activity and related components of Rhodobryum giganteum Par. Phytomedicine. 18(2–3):224–229.
- Chen BJ. 1963. Genera muscorum sinicorum (pars prima). Beijing: Science Press. p. 250–280.
- Darling ACE, Mau B, Blattner FR, Perna NT. 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14(7):1394–1403.
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59(3):307–321.
- Harris ESJ. 2008. Ethnobryology: traditional uses and folk classification of bryophytes. Bryologist. 111(2):169–217.
- Harris ESJ, Yang BL. 2009. Variation and standardization in the use of a Chinese medicinal moss. Econ Bot. 63(2):190–203.
- Hu Y, Guo DH, Liu P, Rahman K, Wang DX, Wang B. 2009. Antioxidant effects of a Rhodobryum roseum extract and its active components in isoproterenol-induced myocardial injury in rats and cardiac myocytes against oxidative stress-triggered damage. Pharmazie. 64(1):53–57.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Li XJ. 1990. Xin Hua Ben Cao Gang Yao. (in Chinese) [Xinhua herbal outline] Vol. 3. Shanghai: Shanghai Scientific and Technical Publishers.
- Li JL, Wang S, Yu J, Wang L, Zhou SL. 2013. A modified CTAB protocol for plant DNA extraction. Bull Bot. 48(1):72–78.
- Li L, Zhao JC. 2009. Determination of the volatile composition of Rhodobryum giganteum (Schwaegr.) Par. (Bryaceae) using solid-phase microextraction and gas chromatography/mass spectrometry (GC/MS). Molecules. 14(6):2195–2201.
- Limpricht KG. 1892. Die laubmoose deutschlands, oesterreichs und der schweiz. Eduard Kummer Leipzig. 2(20):444.
- Minh BQ, Nguyen MAT, von Haeseler A. 2013. Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol. 30(5):1188–1195.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Shi S, Li SL, Zhang SJ, Shen FJ, Niu JY, Li L, Zhao JC. 2021. The complete chloroplast genome of Mnium marginatum (With.) P. Beauv. Mitochondrial DNA Part B. 6(3):837–839.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255.
- Zhang D, Gao FL, Jakovlić I, Zou H, Zhang J, Li WX, Wang JT. 2020. PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol Ecol Resour. 20(1):348–355.