Abstract
Primula calliantha Franch. is an alpine species with an ornamental value that is endemic to the Hengduan Mountains of southwest China. Here, we sequenced and assembled its plastid complete genome, which is a circular molecule of 151,954 bp and contains a large single-copy (83,820 bp) and a small single-copy (17,814 bp) region, separated by a pair of inverted repeats (25,160 bp). There are 131 genes, 86 protein-coding, 37 tRNAs, and 8 rRNAs in the plastome, of which 114 genes, 80 CDSs, 30 tRNAs, and 4 rRNAs are unique, respectively. The P. calliantha plastid genome shows a high level of synteny with its close relatives, P. chionantha, P. purdomii, and P. woodwardii. Phylogenetic analysis based on 60 complete chloroplast genomes of Primulaceae confirmed its delimitation in Primula.
Primula calliantha is a species of Primula classified in section Crystallophlomis Rupr. of the Primulaceae (Hu and Kelso Citation1996). It is endemic to the Hengduan Mountains biodiversity hotspot of southwest China, growing on pastures and meadows among Rhododendron or on rocks of steep humus-clad slopes at elevations 3700–4500 m (Hu and Kelso Citation1996). This alpine endemic species is specifically distributed in the Cang-Shan Mountain of Yunnan Dali, but extends to the Shangri-La country and southeast Xizhang (Richards Citation2003). The flower of P. calliantha has 1–4 umbels, each with three to many purplish violet corolla (Hu and Kelso Citation1996). This species therefore exhibits high ornamental value. To provide scientific basis for the rational utilization and conservation of P. calliantha, we sequenced and assembled the complete plastid genome.
Fresh leaves of P. calliantha were collected from wild plants in Dali County, Yunnan (N 25.67°, E 100.09°) and the corresponding voucher specimen was deposited in the Herbarium of Yunnan Normal University under the accession number of HY202013 (Kunming, China; Prof. Yonghong Zhang, [email protected]). Total genomic DNA was extracted from the isolated chloroplasts using a modified CTAB method (Porebski et al. Citation1997). According to the criterion, we fragmented the DNA and used Illumina Hiseq X Ten sequencer to construct the genomic library for Illumina paired-end (PE) sequencing. A total of 29,141,918 filtered reads were retrieved and used to assemble the plastid genome with NOVOPlasty v4.3.1 (Dierckxsens et al. Citation2017). Using P. chrysochlora Balf. F. et Ward as the reference genome (GenBank accession No: NC_034678), the assembled plastid genome was annotated using GeSeq (Tillich et al. Citation2017).
The complete plastid genome of P. calliantha is a circular molecule with a length of 151,954 bp, with an average coverage of 430, and GC content of 37.0%, respectively. The NCBI accession number of the plastid genome is MZ054238, and the Illumina sequence data are deposited in the NCBI SRA database under accession number SRR14454273. The plastid genome of this species has a large single-copy region of 83,820 bp and a small single-copy region of 17,814 bp, which are separated by a pair of inverted repeats of 25,160 bp. In total, the plastid genome has 131 genes, 86 protein-coding (CDS), 37 tRNAs, and 8 rRNAs are annotated in the plastome, of which 114 genes, 80 CDSs, 30 tRNAs, and four rRNAs are unique, respectively. The gene content, synteny of plastid genome was similar between P. calliantha and its close relatives, P. chionantha, P. purdomii, and P. woodwardii, but slightly differs in plastome length. Moreover, compared with P. calliantha and P. woodwardii, P. chionantha and P. purdomii lack the rp12 gene (Ren et al. Citation2018).
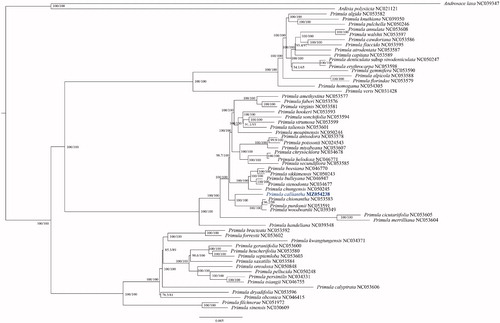
To determine the phylogenetic position of P. calliantha within the genus Primula, 57 Primula sequences were downloaded as well as two Androsace species to serve as outgroups from GenBank. The plastid sequences were aligned using MAFFT (Katoh and Standely Citation2013), and a maximum-likelihood (ML) tree was constructed using IQ-TREE-2 (Minh et al. Citation2020; Katoh and Standley Citation2013). The best fitted model according to the Bayesian information criterion was TVM + F+R10 using ModelFinder (Kalyaanamoorthy et al. Citation2017). The branch support values were tested using the ultrafast bootstrap (UFBoot) setting (Hoang et al. Citation2018) and SH-like approximate likelihood ratio test (SH-aLRT) (Guindon et al. Citation2010) with 10,000 replicates (). The ML tree showed that the 58 Primula species analyzed in this study form a robust monophyletic clade, and P. calliantha is fully resolved in a clade with four species classified to Primula section Crystallophlomis, and is sister to P. chionantha, P. purdomii, P. woodwardia. In contrast, a previous barcode phylogenetic analysis (Yan et al. Citation2015) based on rbcL, matK, and ITS sequence data revealed that P. calliantha is sister to P. boreiocalliantha, and a clade consist of these two species is sister to P. agleniana. However, plastomes of P. boreiocalliantha and P. agleniana are not available, and our research cannot confirm this relationship. Topological structures shown by our phylogenetic tree are generally in agreement with previously phylogenetic studies of Primula with some inconsistencies (Conti et al. Citation2000; Mast et al. Citation2001; Kovtonyuk and Goncharov Citation2009; Yan et al. Citation2015; Ren et al. Citation2018). For example, previous study revealed that sect. Proliferae Pax (Ren et al. Citation2018) and sect. Aleuritia (Conti et al. Citation2000) are monophyletic. But our results with more or different species sampled for these sections revealed they are not monophyletic, indicating that lack of samples affect the results of the phylogenetic analysis. Molecular phylogenetic studies of Primula so far only sampled a small fraction of species in this genus, or used few DNA marks and therefore cannot provide sufficient resolution (Conti et al. Citation2000; Mast et al. Citation2001; Kovtonyuk and Goncharov Citation2009; Ren et al. Citation2018), it is therefore arbitrary to discuss intra-generic systems (e.g. subgenus, section) of Primula. Nevertheless, all these studies indicated that Primula is a robust monophyletic clade, but most sections of Primula are not monophyletic, all of the relationships revealed by molecular evidence between Primula subgenus, sections, and species generally do not correlate with any Primula systems based on morphology (Richards 1993; Mast et al. Citation2001).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Date availability statement
The data that support the findings of this study are available in NCBI at https://www.ncbi.nlm.nih.gov/, reference number [MZ054238]. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA727726 SRR14454273 and SAMN19030661, respectively.
Additional information
Funding
References
- Conti E, Suring E, Boyd D, Jorgensen J, Grant J, Kelso S. 2000. Phylogenetic relationships and character evolution in Primula L., the usefulness of ITS sequence data. Plant Biosyst. 134(3):385–392.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: denovo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18.
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59(3):307–321.
- Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. 2018. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol. 35(2):518–522.
- Hu Q, Kelso S. 1996. Primulaceae. In: Wu CY, Raven PH, editors. Flora of China. Vol. 15. Beijing and St. Louis: Science Press and Missouri Botanical Garden Press; p. 185–188.
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kovtonyuk NK, Goncharov AA. 2009. Phylogenetic relationships in the genus Primula L. (Primulaceae) inferred from the ITS region sequences of nuclear rDNA. Russ J Genet. 45(6):663–670.
- Mast AR, Kelso S, Richards AJ, Lang DJ, Feller DM, Conti E. 2001. Phylogenetic relationships in Primula L. and related genera (Primulaceae) based on noncoding chloroplast DNA. Int J Plant Sci. 162(6):1381–1400.
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, Von Haeseler A, Lanfear R. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 37(5):1530–1534.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Porebski S, Bailey LG, Baum BR. 1997. Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol Biol Rep. 15(1):8–15.
- Ren T, Yang YC, Zhou T, Liu ZL. 2018. Comparative plastid genomes of Primula species: sequence divergence and phylogenetic relationships. Int J Mol Sci. 19(4):1040.
- Richards J. 2003. Primula. 2nd ed. London: Batsford; p. 36–153.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq - versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Yan HF, Liu YJ, Xie XF, Zhang CY, Hu CM, Hao G, Ge XJ. 2015. DNA barcoding evaluation and its taxonomic implications in the species-rich genus Primula L. in China. PLoS One. 10(4):e0122903.