Abstract
Medicago ruthenica is an important perennial forage with multiple characteristics of resistance. In this study, we sequenced and characterized the complete chloroplast genome of M. ruthenica ‘Taihang’, which is 124, 254 bp in length. A total of 108 genes were identified, including 74 protein-coding, 30 tRNA, and four rRNA genes. Phylogenetic analysis based on 27 chloroplast genomes showed that M. ruthenica ‘Taihang’ has a close relationship with M. ruthenica from Qinghai Province, China. The data are useful in better understanding the genetic diversity and stress resistance of Medicago and contribute to the phylogenetic study of Trifolieae.
Medicago L. is classified in the Fabaceae and is an important forage plant because of its rich nutrition and digestibility (Choi et al. Citation2019). Medicago ruthenica, with the characteristics of cold resistance, drought resistance, salt and alkali resistance, infertility resistance (Shu et al. Citation2018), is equipped with wide adaptability which makes the morphology vary greatly under different environmental conditions (Cui et al. Citation1998). In China, three cultivars have been described as cultivars of M. ruthenica, including ‘Tumote’, ‘Zhi Lixing’ and ‘Taihang’. The cultivar ‘Taihang’, which has a large number of leaves, good quality, high grass yield and seed yield, is more suitable for planting in northern semi-arid areas in China and other similar conditions. To date, several barcode analyses have been performed using matK and trnH-psbA (Nadia et al. Citation2014; Badr et al. Citation2020), and a complete chloroplast genome of M. ruthenica from Qinghai Province, China was assembled and published previously (Xie et al. Citation2021). In this research, we aimed to establish and characterized the chloroplast (cp) genome of the ‘Taihang’ cultivar to provide additional genomic data for the phylogenetic study of this species and the Trifolieae.
In this study, fresh leaves of M. ruthenica ‘Taihang’ were collected from experimental plots of the Shanxi Agricultural University of China, located at 112.5824 E, 37.4183 N. The voucher specimen is stored in the herbarium of Shanxi Agricultural University (Specimen No. SXAU-CoGS-20Mr01, Hua Zhong, [email protected]). Chloroplasts were isolated from 5 g of fresh leaves by gradient centrifugation on Percoll. Then, the chloroplast DNA was extracted using the CTAB method (Doyle JJ and Doyle JL Citation1987). A 350 bp DNA library was constructed and PE-150 bp reads were sequenced on an Illumina Hiseq 2500 platform. To filter reads from the chloroplast genome, approximately 348.4 Mb of trimmed high-quality reads were mapped to the previously published cp genomes (NC_042841.1, NC_042849.1, NC_042854.1, NC_003119.8 and NC_032066.1) Minimap2 v2.13-r850 with default parameters (Li Citation2018). The resulting aligned reads were co-assembled using GetOrganelle v1.7.3.1 (Jin et al. Citation2020), and one circular contig was produced. Genome annotation was conducted using the online tool GeSeq (https://chlorobox.mpimp-golm.mpg.de/geseq.html, Tillich et al. Citation2017) and manually corrected.
The complete chloroplast genome of M. ruthenica ‘Taihang’ (GenBank accession number: MW703984.1) has a circular chromosome that is 124,254 bp in length with a GC content of 34.29%. A total of 108 genes were annotated, including 74 protein-coding, 30 tRNA, and four rRNA genes. Fifteen of the genes contain one intron and one of them contains two introns. Compared with the chloroplast genomes previously published about the other plants of Medicago L. (Choi et al. Citation2019; Zhao et al. Citation2021), M. ruthenica (MH901635.1) published by Xie et al. (Citation2021) lacks the gene ndhD which encodes NADH dehydrogenase subunit 4, and M. ruthenica ‘Taihang’ (MW703984.1) lacks the gene petN which encodes cytochrome b6/f complex subunit VIII. In addition, the gene sequence from clpP to psaA is reversed in M. ruthenica MH901635.1 and MW703984.1. The genes trnC-GCA and rpoB in MH901635.1 are located between rpoC1 and psaB, while they are located between psbM and rpoC1 in MW703984.1 and which is similar to the other plants of Medicago L. (Choi et al. Citation2019). However, the chloroplast genome of two cultivars still shows a high level of gene synteny.
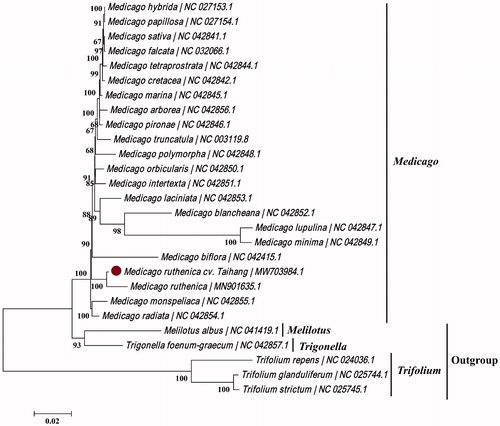
To reveal the phylogenetic position of M. ruthenica ‘Taihang’, a maximum-likelihood (ML) tree was constructed using 27 complete chloroplast genome sequences, including 22 Medicago species, three Trifolium species, and Melilotus albus and Trigonella foenum-graecum serving as outgroup taxa. All sequences were downloaded from GenBank and were aligned using MAFFT v7.311 using the default settings (Katoh and Standley Citation2013). The ML tree was produced by MEGA7 v7.0.26 (Kumar et al. Citation2016) using 1,000 bootstrap replicates and the Tamura-Nei model + Uniform rates nucleotide substitution model. The phylogenetic tree showed that M. ruthenica ‘Taihang’ was fully resolved on a branch with M. ruthenica (MH901635.1) within a monophyletic clade with other species classified to the genus Medicago (). In addition, barcode analyses had also been carried out using genes matK and trnH-psbA, and which presented the same relationship between ‘Taihang’ and other Medicago L. plants. The complete analysis of the chloroplast genome of M. ruthenica ‘Taihang’ is useful for studying the genetic diversity and stress resistance of Medicago plants.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The raw data that support the findings of this study is openly available in NCBI Sequence Read Archive at [https://www.ncbi.nlm.nih.gov/sra/SRR14114639] under the BioProject ID PRJNA718640 and the annotated chloroplast genome has been deposited in Genbank [https://www.ncbi.nlm.nih.gov/nuccore/MW703984.1/] under the reference number MW703984.1.
Additional information
Funding
References
- Badr A, El-Sherif N, Aly S, Ibrahim SD, Ibrahim M. 2020. Genetic diversity among selected Medicago sativa cultivars using inter-retrotransposon-amplified polymorphism, chloroplast DNA barcodes and morpho-agronomic trait analyses. Plants. 9(8):995.
- Choi IS, Jansen R, Ruhlman T. 2019. Lost and found: return of the inverted repeat in the legume clade defined by its absence. Genome Biol Evol. 11(4):1321–1333.
- Cui HB, Wei Z, Huang YZ. 1998. Medicago ruthenica (L.) Trautv. Flora of China. Vol. 42. Beijing, China: Beijing Science Press; p. 318.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Jin J-J, Yu W-B, Yang J-B, Song Y, dePamphilis CW, Yi T-S, Li D-Z. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Li H. 2018. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 34(18):3094–3100.
- Nadia Z, Maroua G, Hela BR, Houda CK, Abdelmajid H, Neila TF, Sonia M. 2014. Evolutionary and demographic history among Maghrebian Medicago species (Fabaceae) based on the nucleotide sequences of the chloroplast DNA barcode trnH-psbA. Biochem Syst Ecol. 55:296–304.
- Shu Y, Li W, Zhao J, Liu Y, Guo C. 2018. Transcriptome sequencing and expression profiling of genes involved in the response to abiotic stress in Medicago ruthenica. Genet Mol Biol. 41(3):638–648.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq - versatile and accurate annotation of organelle genomes . Nucleic Acids Res. 45(W1):W6–W11.
- Xie J, Mao J, Li Z, Liang C, Ou W, Tang J, Liu H, Wang H, Ji Z, Kazhoucairang , Shen YF. 2021. Complete chloroplast genome of a high-quality forage in north China, Medicago ruthenica (Fabaceae: Trifolieae). Mitochondrial DNA Part B. 6(1):29–30.
- Zhao Y, Zeng W, Li W, Bi Y. 2021. Complete chloroplast genome sequence of the drought and heat-resistant Chinese Alfalfa Landrace, Medicago Sativa ‘Deqin’. Mitochondrial DNA B Resour. 6(4):1488–1489.