Abstract
In this study, we assembled a complete mitochondrial genome of Codonobdella sp. sample from Lake Baikal, Russia and reassembled third party raw data of Piscicola geometra. Mitogenomes of both freshwater piscine leeches consist of 37 genes, including 13 protein-coding genes (PCGs), two ribosomal genes (12S and 16S), 22 transport RNA genes, and one control region. The complete mitogenome of Codonobdella sp. is 14,486 bp long and A + T biased (75.51%). The complete mitogenome of Piscicola geometra is 14,788 bp long and A + T biased (78.27%).
The Codonobdella sp. is a potentially new species of abyssal piscine leeches belonging to the genus endemic to Lake Baikal, which, according to V. Epstein (Citation1987), was represented by the only species C. truncata Grube, 1873, morphologically and ecologically different from the studied sample. Codonobdella sp. was primary found by I. Kaygorodova (Citation2012) on amphipods throughout the lake on depth 40–860 m. It differs from the C. truncata at least by existence of a distinctive pigmentation on the dorsal side, size and shape of body. The further morphological analysis is necessary to describe the new species. Decoding the complete mitochondrial genome may serve as a foundation for evolutionary investigations. The study of genomic sequences may shed light on nuances of molecular evolution mechanisms as well as conditions of endemic species existence and survival.
The samples of Codonobdella sp. were taken from fishing nets in the Maloe More Strait of Lake Baikal, Russia: 53°15'36.9"N 107°06'45.7"E. Biological material is deposited in Limnological Institute, Russia (contact person Dr. Irina Kaygorodova, [email protected]) under voucher no. B45. The total DNA was extracted with the DiaGene Cell Culture kit. The reads were obtained through Illumina NextSeq 550 at the IC&G Center of Genomic Investigations (Novosibirsk, Russia). Due to incompleteness of the Piscicola geometra (L., 1761) MT628553 assembly, we used raw SRR12518725 reads from the Short Read Achieve (SRA) to reassemble its mitogenome. This widespread freshwater species is the closest Piscicolidae relative, available in public sources. The assembly of both samples was performed in MIRA5 (Chevreux et al. Citation1999). Protein annotation was carried out by BLASTp search of translated open reading frames. Transfer RNAs were predicted using ARAGORN (Laslett and Canback Citation2004).
The complete mitogenomes of Codonobdella sp. and Piscicola geometra are 14,486 and 14,788 bp long with the AT content of 75.51% and 78.27%, correspondingly. A total of 37 genes were identified on both mitogenome sequences, including 13 PCGs, two ribosomal genes (12S and 16S), 22 transport RNA genes, and one putative control region. The genes order is the same as for marine leech Zeylanicobdella arugamensis de Silva, 1963 (KY474378), the only complete mitogenome of the Piscicolidae family available in GenBank prior to this study.
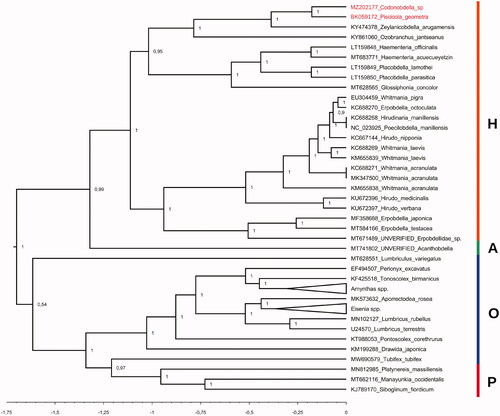
Phylogenetic position of the first obtained mitogenome sequence of Codonobdella sp. relative to 57 evolutionarily close taxa (Oligochaeta, Hirudinea and Polychaeta) available in the NCBI database was inferred using BEAST v2.6.0 (Bouckaert et al. Citation2019) under optimal substitution model (GTR + G + I) selected by jModelTest v2.1.10 (Darriba et al. Citation2012). The resulting tree () showed that all samples grouped into distinct clades according to their taxonomy. The Codonobdella sp. and P. geometra sequences, together with that of Z. arugamensis, formed the clade of the Piscicolidae representatives within the lineage of Rhynchobdellida. This is fully consistent with the modern classification of leech taxa.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The original data of this study have open access at https://www.ncbi.nlm.nih.gov/, reference number MZ202177. The associated BioProject is accessible at https://www.ncbi.nlm.nih.gov/sra/PRJNA733851, with SRA SRR14709013 and BioSample SAMN19460315. The Third Party Annotation is deposited at the DDBJ/ENA/GenBank databases under the accession number TPA: BK059172.
Additional information
Funding
References
- Bouckaert R, Vaughan TG, Barido-Sottani J, Duchêne S, Fourment M, Gavryushkina A, Heled J, Jones G, Kühnert D, De Maio N, et al. 2019. BEAST 2.5: An advanced software platform for Bayesian evolutionary analysis. PLoS Comput Biol. 15(4):e1006650.
- Chevreux B, Wetter T, Suhai S. 1999. Genome sequence assembly using trace signals and additional sequence information. Computer Science and Biology: Proceedings of the German Conference on Bioinformatics. 99, p. 45–56. http://citeseerx.ist.psu.edu/viewdoc/download? doi=10.1.1.23.7465&rep=rep1&type=pdf
- Darriba D, Taboada G, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9(8):772.
- Epstein V. 1987. Annelida. In: Scarlato O.A. (ed.). Key of the freshwater fish parasites of the USSR Fauna. Leningrad: Nauka 3(2): p. 340–372.
- Kaygorodova I. 2012. A revised checklist of the Lake Baikal Hirudinida fauna. Lauterbornia. 75:49–62.
- Laslett D, Canback B. 2004. ARAGORN, a program for the detection of transfer RNA and transfer-messenger RNA genes in nucleotide sequences. Nucleic Acids Res. 32(1):11–16.