Abstract
Abies forrestii is endemic to southwest China and ecologically important as a major component of the cold temperate forests. This study was the first report complete chloroplast (cp) genome of A. forrestii. The complete chloroplast genome was 120,022 bp in size. In total, 114 genes were identified, including 68 peptide-encoding genes, 35 tRNA genes, four rRNA genes, six open reading frames and one pseudogene. Thirteen genes contain introns. In phylogenetic analysis, A. forrestii was found to be closely related with A. nukiangensis, A. fanjingshanensis and A. delavayi subsp. fansipanensis. Our study will provide potential genetic resources for further evolutionary studies of this ecologically important species.
The genus Abies Miller is the second largest one in the family Pinaceae, consisting of approximately 48 species (Liu Citation1971). It has a discontinuous distribution in eastern Asia, eastern and western North America, and the Mediterranean basin (extending into Europe and southwest Asia) (Farjon Citation1990, Citation2001). Abies forrestii Coltm.-Rog. is endemic to southwest China, including northwest of Yunnan province, southwest of Sichuan province and east Tibet. This fir species is ecologically important as a major component of the cold temperate forests and provide a basic home for a great diversity of animals and plants (Liu Citation1971). Besides, they are also suitable materials for performing conservation, ecological and evolutionary studies (Xiang et al. Citation2015). Here, we assembled and characterized the complete plastome of A. forrestii. It will provide potential genetic resources for further relevant evolutionary studies.
The plant material of Abies forrestii was collected from a single individual that lives in Daofu county, Sichuan province (30.79°N, 101.29°E). Voucher specimen and DNA sample (Zhang X.-C., No. 5830) were deposited in the herbarium of Institute of Botany, CAS (PE) (http://pe.ibcas.ac.cn/, Qin Ban, [email protected]). Total genome DNA was extracted with the Ezup plant genomic DNA prep kit (Sangon Biotech, Shanghai, China). Total DNA was used to generate libraries with an average insert size of 350 bp, which were sequenced using the Illumina HiSeq X platform. In total, ca. 10.2 million high-quality clean reads (150 bp PE read length) were generated with adaptors trimmed. The CLC de novo assembler (CLC Bio, Aarhus, Denmark), BLAST, GeSeq (https://chlorobox.mpimp-golm.mpg.de/geseq.html) (Tillich et al. Citation2017), and tRNAscan-SE v1.3.1 (Schattner et al. Citation2005) were used to align, assemble, and annotate the plastome. Genome annotation was performed by comparing the sequences with the cp genomes of Abies koreana (KP742350) and A. neprolepis (KT834974).
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at [https://www.ncbi.nlm.nih.gov] under the accession no. MH706715. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA747743, SRP328853 and SAMN20294543 respectively. The full length of Abies forrestii chloroplast genome was 120,022 bp. The chloroplast genome showed a typical quadripartite structure that consisted of a pair of IR regions (264 bp) separated by the LSC (66,363 bp) and SSC (53,131 bp) regions, which was similar to the majority of cp genomes in Pinaceae. The GC content were 38.30%. A total of 114 genes were contained in the cp genome (68 peptide-encoding genes, 35 tRNA genes, four rRNA genes, six open reading frames and one pseudogene). Fifty-three protein coding, 16 tRNA genes, three open reading frames and one pseudogene are located in the LSC region, while 15 protein-coding, 17 tRNA genes, 4 rRNA and 3 open reading frames are located in the SSC region, respectively. Only one tRNA gene (trnI-CAU) is duplicated and located on the IR regions. All ndh genes have been lost in the genome of A. forrestii like other cp genomes of family Pinaceae. Among the protein-coding genes, two genes (rps12 and ycf3) contained two introns, and other eleven genes (trnK-UUU, trnV-UAC, rpoC1, atpF, trnG-GCC, petB, petD, rpl16, rpl2, trnL-UAA, trnA-UGC) had one intron each. In previous studies, short inverted repeat sequences which consist of trnS-psaM-ycf12-trnG and trnG-ycf12-psaM-trnS (1183 bp) are located in 52-kb inversion points of the cp genome of A. forrestii. Length and sequence of inverted repeats from A. forrestii is identical with those of A. koreana (Yi et al. Citation2015).
Twenty-one published chloroplast genomes were selected to infer the phylogenetic relationships among 16 different fir species with Keteleeria davidiana as the outgroup (Shao et al. Citation2018, Citation2019; Fu et al. Citation2019; Su et al. Citation2019; Wu et al. Citation2019; Li et al. Citation2020; Zhang et al. Citation2020). These sequences were fully aligned with MAFFT v7.3 (Suita, Osaka, Japan) (Katoh and Standley Citation2013). For conducting Maximum Likelihood (ML) analyses, the maximum likelihood (ML) inference was performed using GTRþIþC model with RAxML v.8.2.1 (Karlsruhe, Germany) (Stamatakis Citation2014) on the CIPRES cluster service (Miller et al. Citation2010). MrBayes was run for 1,000,000 generations, sampling and printing every 100 generations. Based on twenty-two cp genome sequences, these Abies species are supported as one monophyletic lineage with extremely high probabilities (BSML = 100) (). And A. forrestii was closely related with A. nukiangensis, A. fanjingshanensis and A. delavayi subsp. fansipanensis ().
Figure 1. Phylogram of Abies forrestii obtained from the maximum likelihood analysis of the whole chloroplast genome sequences. Numbers on branches are support values [maximum likelihood bootstrap values (BSML).
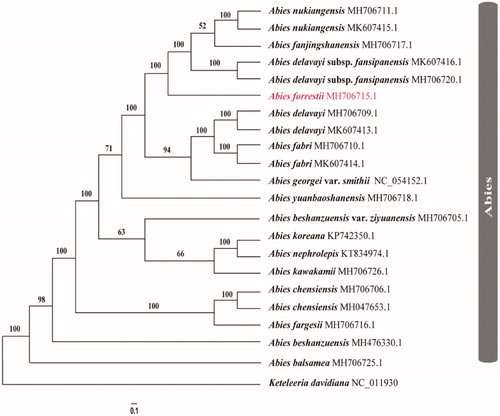
This study provides new insight into the cp genome evolution and phylogenetic relationships of Abies forrestii. Moreover, it would be fundamental to formulate potential conservation and management strategies for this ecologically important species of southwest China.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MH706715. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA747743, SRP328853, and SAMN20294543, respectively.
Additional information
Funding
References
- Farjon A. 1990. Pinaceae: drawings and descriptions of the genera Abies, Cedrus, Pseudolarix, Keteleeria, Nothotsuga, Tsuga, Cathaya, Pseudotsuga, Larix and Picea (Regnum Vegetabile 121). Königstein: Koeltz Scientific Books; p. 9–109.
- Farjon A. 2001. World checklist and bibliography of conifers. 2nd ed. London: The Royal Botanic Gardens, Kew.
- Fu ZX, Wang XY, Fan PZ, Tian XY, Shao YZ. 2019. The complete chloroplast genome of the endangered Pinaceae species Abies ziyuanensis and its phylogenetic implications. Mitochondrial DNA B. 4(1):137–138.
- Li JR, Chen JR, Wang JK, Zheng WL. 2020. The complete chloroplast genome of Abies georgei Orr var. smithii, a species endemic to the Qinghai-Tibet Plateau, China. Mitochondrial DNA B. 53(3):2553–2554.
- Liu TS. 1971. A monograph of the genus Abies. Taipei (Taiwan): Department of Forestry, College of Agriculture, National Taiwan University.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. 2010 Gateway Computing Environments Workshop (GCE 2010). IEEE Computer Society; New Orleans, LA.
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33(Web Server issue):W686–W689.
- Shao YZ, Chen Y, Lu XF, Ye YZ, Yuan ZL. 2019. Next-generation sequencing yields the complete chloroplast genome of Abies kawakamii. Mitochondrial DNA B Resour. 4(1):29–30.
- Shao YZ, Hu JT, Fan PZ, Liu YY, Wang YH. 2018. The complete chloroplast genome sequence of Abies beshanzuensis, a highly endangered fir species from south China. Mitochondrial DNA B. 3(2):923–924.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Su L, Zhao PF, Lu XF, Shao YZ. 2019. The complete chloroplast genome sequence of Abies chensiensis (Pinaceae). Mitochondrial DNA B Resour. 4(2):3262–3263.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq - versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Wu S, Lu XF, Zhao PF, Shao YZ. 2019. Next-generation sequencing yields the complete chloroplast genome of Abies balsamea. Mitochondrial DNA B. 4(1):1445–1446.
- Xiang QP, Wei R, Shao YZ, Yang ZY, Wang XQ, Zhang XC. 2015. Phylogenetic relationships, possible ancient hybridization, and biogeographic history of Abies (Pinaceae) based on data from nuclear, plastid, and mitochondrial genomes. Mol Phylogenet Evol. 82 Pt A:1–14.
- Yi DK, Yang JC, So S, Joo MJ, Kim DK, Shin CH, Lee YM, Choi K. 2015. The complete plastid genome sequence of Abies koreana (Pinaceae: Abietoideae). Mitochondrial DNA. 27(4):1.
- Zhang YY, Xiang RC, Dong NL, Liu YY, Shao YZ. 2020. Next-generation sequencing yields the complete chloroplast genome of Abies yuanbaoshanensis, an endangered species from South China. Mitochondrial DNA B. 5(4):3821–3822.
