Abstract
The structure and composition of the mitogenome of a bivalve mollusk, denoted as Mactra sp. (MT780813), has been obtained. The genome has a variable organization: it includes 12 protein-coding genes, 28 tRNAs, and two rRNA genes. Its content is sufficiently different from that of nearest specimen, accessed from GenBank, supposedly belonging to the other gender. All genes are encoded on the “+”-strand. All protein-coding genes are initiated with ATG codon. Analysis confirms the close topological position of the GenBank Mactra chinensis (KJ754823) and our M. sp. specimen on gene tree. Above data suggesting female- vs. male-type mitogenomes or cryptic species presence.
Introduction
Bivalve mollusk the Chinese surf clam, Mactra chinensis Philippi, 1846, is one of the commercially valuable species of the family Mactridae (Bivalvia: Venerida; WoRMS, http://www.marinespecies.org/aphia.php?p=taxdetails&id=367847; Ni et al. Citation2012; Shen et al. Citation2014). The species is a typical member of benthic communities distributed in boreal nearshore sandy habitats, usually at a depth of 7–10 m (Scarlato Citation1976). Its range extends to the West Pacific: from the Kuril Islands to coastal waters of the Sea of Japan and off the Korean Peninsula down to the Yellow Sea (Scarlato Citation1976). Mactra chinensis has appeared to vary in shell color pattern between the southern and northern parts of the species range: in China and South Korea waters, its shells are colored yellowish, without brown rays, while in the Russian part of the Sea of Japan, shells have a typical brown-and-grayish pattern with brown and violet purple rays (Scarlato Citation1976; Lutaenko and Noseworthy Citation2012). Bivalve mollusks of the order Venerida, including M. chinensis and many other members of Bivalvia, were subjected to a comparative molecular phylogenetic analysis by sequences of certain mitochondrial genes, nuclear loci, and complete mitogenome (Theologidis et al. Citation2008; Ni et al. Citation2012; Shen et al. Citation2014; Fernández-Pérez et al. Citation2017; Reunov et al. Citation2019). Several studies have revealed a number of taxonomic issues at the species, genus, and family levels. Mitogenome has been described for many bivalve species and appeared to be very variable in size, structure, and even order of genes (Yuan et al. Citation2013; Shen et al. Citation2014; Fernández-Pérez et al. Citation2017). Many bivalves show a phenomenon of double uniparental inheritance (DUI) of single mtDNA genes and whole mitogenome in females (F) and males (M) causing their drastic divergence (Zouros et al. Citation1994; Skibinski et al. Citation1994; Theologidis et al. Citation2008; Fernández-Pérez et al. Citation2017 etc.).
The aim of the present work was to study the mitogenome of one Mactra specimen, whose shell originally was not identified exactly and was referred to as Mactra sp., and also to clarify its species name, investigate the mitogenome composition of this organism, and show its place among a few relatives from the genera Mactra, Colomactra, and Lutraria using literature sources.
Materials and methods
The mitogenome of Mactra sp. (GenBank accession no. MT780813, voucher no. WB-F) was obtained by Illumina sequencing technology (Novaseq 6000, USA) and assembled in SPAdes v. 3.11.0. (Bankevich et al. Citation2012) using the adductor muscle specimen from the sampled individual in Shahekou District, Dalian, China (sex was not determined for this mollusk due to empty gonads at the sampling time). The original mitogenome annotation was obtained from a sequencing laboratory: Huitong Biotechnology, Shenzhen, China. The voucher specimen was collected by the College of Fisheries and Life Science, Dalian Ocean University, #52 Heishijiao Street, Shahekou District, Dalian, Liaoning 116023, China.
The produced consensus sequence of the complete genome was annotated using the MITOS Web Server (Bernt et al. Citation2013). The mitogenome map was built with GeSeq software (Tillich et al. Citation2017). The nucleotide sequences were aligned by Muscle or ClustalW for the whole mitogenome or separately for protein-coding genes (PCGs) as implemented in MEGA-X (Kumar et al. Citation2018). At the next step, two substitution models, TrN + G + I or HKY + G + I, were determined as best fit and used for neighbor-joining, maximum likelihood, and maximum parsimony (NJ, ML, and MP, respectively) reconstructions of gene trees for phylogenetic analysis; these two models were applied, respectively, for NJ and ML. For building gene trees, the MEGA-X software was used. For estimating the node’s support, the default settings and bootstrapping mode with n = 1000 replications were applied. NJ () and other trees (not shown) were built on 12 PCGs sequences of seven mitochondrial genomes. The outgroup, where necessary, was Lutraria whose representatives belong to a different subfamily.
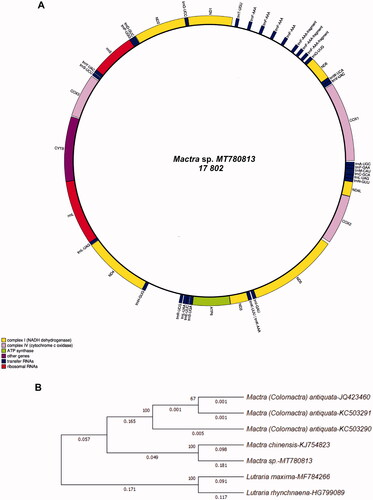
Figure 1. The organization of Mactra sp. MT780813 (Mactridae) mitochondrial genome and the position of the specimen on the gene tree with the nearest relatives. (A) All genes are encoded on the same strand (+). The transfer RNA genes are designated by three-letter amino acid codes. The rRNA subunits, 12S rRNA and 16S rRNA, are shown in this annotation as rRNA-S (rrnS, within a circle) and rRNA-L (rrnL, within a circle), respectively. (B) Original NJ gene tree demonstrating the relationships among the nearest relatives of Mactra sp. (MT780813 sequence), built on the basis of 12 PCGs sequences (no ATP8 identified in our sequence, as implemented in and in Supplement) of seven mitochondrial genomes. Numerals at the nodes indicate support values for tree topology based on 1000 replicates of the bootstrap test. Numerals below the branches indicate the composition bias calculated for seven sequences in the analysis. Congruent tree topology was obtained for NJ, ML, and MP-tree reconstructions (not shown).
Results and discussion
The mitogenome annotation of the Mactra sp. MT780813 specimen was characterized as circular molecular with obvious deviations from the mitochondrial gene order typical of vertebrates and depauperate in this and some other features from the closest specimen in the GenBank (https://www.ncbi.nlm.nih.gov/), as shown by BLAST searches on 27 June 2020 and 22 August 2020. It was found belonging to M. chinensis KJ754823 (identified as female’s mitogenome, F; Fernández-Pérez et al. Citation2017). MITOS detected the following content for the Mactra sp. MT780813 specimen: 12 PCGs, two D-loops or Control Regions (CR), 12S rRNA, and 16S rRNA, 27 tRNAs including two tRNA-Leu’s, tRNA-Ser’s, and tRNA-Asn’s, and also four tRNA-Phe’s (). No ATPase 8 known for some bivalves (Shen et al. Citation2014) were found in this genome. Each of the four copies of tRNA-Phe was accompanied by the 189 bp non-coding region, hence, representing tandem repeats (). As follows from Supplementary Table 1 data, tRNA gene numbers and NCR, noncoding regions sharply differ in two compared sequences; tRNA gene numbers are 27 for MT780813 (this paper) and 20 for KJ754823 (Shen et al. Citation2014). Three NCR have been listed in our specimen, while no one mentioned KJ754823 (Supplementary Table 1). However, for PCGs the overall diversity is reasonably big too, as illustrated in the paper and indicated by the composition bias calculated for seven sequences in the analysis (). There are also the tandem repeats as noted above.
The mitogenome of Mactra sp. MT780813 was 17,802 bp in length with the above-noted complement of genes and structural elements, which differed from the earlier described mitogenome sequence KJ754823 (Shen et al. Citation2014). The overall base composition was 41.2 ± 0.51% T (the values presented hereafter are mean ± standard error), 13.4 ± 0.51% C, 23.1 ± 0.51% A, and 22.2 ± 0.51% G. By this composition, this mitogenome did not differ from the sequence KJ754823 by Shen et al. (Citation2014), F = 1.015, p > 0.39. All 12 PCGs started with the ATG initiation codon. A few atypical starts and stop codons were also found as well as in many other fish and shellfish taxa (Kartavtsev et al. Citation2007; Shen et al. Citation2014; Li et al. Citation2014; Turanov et al. Citation2019; Zheng et al. Citation2019).
In order to investigate the phylogenetic position of Mactra sp. MT780813 among the nearest relatives, NJ, ML, and MP phylogenetic analyses were carried out on the basis of complete mitogenome sequences from a few nearest relatives from the family Mactridae and two species from another subfamily as an outgroup (). As shown in , Mactra sp. MT780813 is more closely related to the specimen M. chinensis KJ754823. However, the two Mactra sequences potentially have large distance scores (e.g. 0.181 for composition bias), in accordance with the pattern provided in the NJ tree (). Congruent tree topology was obtained for all three reconstructions, that is, NJ, ML, and MP-trees (the two latter are not shown). The topology for the Mactridae representatives in well matches the wider comparison of F-mitogenome sequences of bivalves (Fernández-Pérez et al. Citation2017). Thus, judging by the known interspecies differences for the other two species representing this genus, with a p-distance score equal to 0.073–0.083 (Shen et al. Citation2014), we can conclude that the specimen of Mactra sp. MT780813 probably belonged to the species M. chinensis of M-type (male genome). The alternative of different sex lines representation in the source specimens due to DUI is differences occurred in the hidden taxa that might be specified as an unknown taxon (Mactra sp.) different from typical M. chinensis. As is well known, M-type is highly divergent from F-type in many bivalve mollusks (Theologidis et al. Citation2008). In several bivalve mollusks, differences between the M- and F-type sequences in p-distance for the 16S rRNA and cyt b genes averaged at 0.366 ± 0.038 and 0.507 ± 0.044, respectively (Theologidis et al. Citation2008). While for the two representatives of the nearest genera of Mactridae, M. chinensis and Colomactra (Mactra) antiquate (MolluscaBase, Citation2020), the p-distance score between M- and F-type reached 0.0953 ± 0.0023 and 0.0949 ± 0.0016, respectively, or even greater depending on sequence assemblage (mitogenome, estimates based on our and GenBank data). Cryptic species among Colomactra (Mactra) antiquata (Ni et al. Citation2012; Yuan et al. Citation2013; Shen et al. Citation2014) and Mactra (Reunov et al. 2019) were considered, but there was no clarification about the DUI point. As noted in the introduction, such bivalve taxa as Mytilidae, Veneridae, etc., that exhibiting the DUI phenomenon of mtDNA inheritance, may have a level of F- vs. M-type intraspecies divergence (Zouros et al. Citation1994; Skibinski et al. Citation1994; Theologidis et al. Citation2008; Fernández-Pérez et al. Citation2017) higher than the divergence typical for different species. The assembly artifacts and Labs’ made mistakes if sequencing made in different conditions may possibly cause such an obscure conclusion. However, in our case the most natural and highly probable are two reasons: (i) the gender differences that have been established earlier for the relatives of these mollusks’ (Theologidis et al. Citation2008; Fernández-Pérez et al. Citation2017) because source tissues (adductor muscle) are same for two compared sequences, MT780813 (this paper) and KJ754823 (Shen et al. Citation2014) or (ii) differences occurred in the hidden taxa that might be specified by Mactra sp. MT780813 vs. M. chinensis KJ754823. By the orthodox viewpoint the adductor specimen from Mactra sp. MT780813 must be F-type. However, the matter is more complicated, as summed up in recent papers (Kyriakou et al., Citation2015; Schwartz Citation2021). So, sometimes, females could get M-type mitochondria. Inversely, male’s repression mechanism with the sex-specific protein-binding site for turn-off M-type in the somatic genome might not act properly leaving the heteroplasmy and F-type presence.
Thus, despite the probability of the existence of cryptic taxa, accurate sex determination, homogenous gender representation, and the comparison of a voucher collection of specimens are the ultimate requirement for phylogenetic reconstructions and taxonomic identification, as exemplified elsewhere (Fernández-Pérez et al. Citation2017), including individual-based assessment of the F- vs. M-type mtDNA difference. This will be the aim of the next research by our team.
Supplemental Material
Download MS Word (25.5 KB)Acknowledgments
The authors thank E. P. Shvetsov for MS proofreading. We also grateful to Dr. S. V. Turanov for help with MS editing at earlier stages.
Disclosure statement
The authors have no conflicts of interest to declare. The authors are solely responsible for the content and the writing of this paper.
Data availability statement
The mitogenome of Mactra sp. can be obtained from GenBank under accession no. MT780813. The voucher specimen was collected and stored by the College of Fisheries and Life Science, Dalian Ocean University, #52, Heishijiao Street, Shahekou District, Dalian, Liaoning 116023, China. It is deposited at this institution, as a specimen with voucher no. WB-F and curated at the Lab by person Nie, Hongtao (Tel. +86-0411-84763083; e-mail: [email protected]). The associated BioProject, SRA, and Bio-Sample numbers are PRJNA745510, SUB9992288, and SAMN20168722, respectively. Also, data from the paper are available on web-page: (13) (PDF) Complete mitochondrial genome of Mactra chinensis (Bivalvia: Macridae) (researchgate.net).
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Fernández-Pérez J, Nantón A, Ruiz-Ruano FJ, Camacho JPM, Méndez J. 2017. First complete female mitochondrial genome in four bivalve species genus Donax and their phylogenetic relationships within the Veneroida order. PLoS One. 12(9):e0184464.
- Kartavtsev YP, Jung S-O, Lee Y-M, Byeon H-K, Lee J-S. 2007. Complete mitochondrial genome of the bullhead torrent catfish, Liobagrus obesus (Siluriformes, Amblycipididae): genome description and phylogenetic considerations inferred from the Cyt b and 16S rRNA genes. Gene. 396(1):13–27.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Kyriakou E, Kravariti L, Vasilopoulos T, Zouros E, Rodakis GC. 2015. A protein binding site in the M mitochondrial genome of Mytilus galloprovincialis may be responsible for its paternal transmission. Gene. 562(1):83–94. doi:https://doi.org/10.1016/j.gene.2015.02.047.
- Li H, Zhang J, Li H, Gao X, He C. 2014. Development and characterization of 13 polymorphic microsatellite markers for the Chinese surf clam (Mactra chinensis) through Illumina paired-end sequencing. Conservation Genet Resour. 6(4):877–879.
- Lutaenko KA, Noseworthy RG. 2012. Catalogue of the living Bivalvia of the continental coast of the Sea of Japan (East Sea). Vladivostok (Russia): Dalnauka; p. 247.
- MolluscaBase. 2020. Mactra chinensis Philippi, 1846. Brussels (Belgium): LifeWatch; Accessed 2020 September 18]. http://www.marinespecies.org/aphia.php?p=taxdetails&id=367847
- Ni L, Li Q, Kong L, Huang S, Li L. 2012. DNA barcoding and phylogeny in the family Mactridae (Bivalvia: Heterodonta): evidence for cryptic species. Biochem Syst Ecol. 44:164–172.
- Reunov AA, Lutaenko KA, Zakharov EV. 2019. Genetic divergence in bivalve mollusk Mactra chinensis related with disproporsional geteromorphism in male gametes. Proc Russ Acad Sci. 455(5):1–4.
- Scarlato OA. 1976. Bivalve mollusks – Bivalvia. In: Animals and plants of Peter the Great Bay. Moscow (Russia): Nauka. p. 95–107.
- Schwartz JH. 2021. Evolution, systematics, and the unnatural history of mitochondrial DNA. Mitochondrial DNA A DNA Mapp Seq Anal. 32(4):126–151. doi:https://doi.org/10.1080/24701394.2021.1899165.
- Shen X, Meng XP, Chu KH, Zhao NN, Tian M, Liang M, Hao J. 2014. Comparative mitogenomic analysis reveals cryptic species: a case study in Mactridae (Mollusca: Bivalvia). Comp Biochem Physiol Part D Genomics Proteomics. 12:1–9.
- Skibinski DOF, Gallagher C, Beynon CM. 1994. Mitochondrial DNA inheritance. Nature. 368(6474):817–818.
- Theologidis I, Fodelianakis S, Gaspar MB, Zouros E. 2008. Doubly uniparental inheritance (DUI) of mitochondrial DNA in Donax trunculus (Bivalvia: Donacidae) and the problem of its sporadic detection in Bivalvia. Evolution. 62(4):959–970.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq - versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Turanov SV, Rutenko OA, Kartavtsev YP. 2019. Complete mitochondrial genome of Stichaeus grigorjewi Herzenstein, 1890 (Zoarcales: Stichaeidae). Mitochondrial DNA Part B. 4(1):899–901.
- Yuan Y, Kong L, Li Q. 2013. Mitogenome evidence for the existence of cryptic species in Coelomactra antiquata. Genes Genom. 35(6):693–701.
- Zheng Y, Chen Y, Chen P. 2019. The complete mitochondrial genome of an eelpout Lycodes ygreknotatus (Teleostei: Zoarcidae). Mitochondrial DNA Part B. 4(1):616–617.
- Zouros E, Ball AO, Saavedra C, Freeman KR. 1994. Mitochondrial DNA inheritance. Nature. 368(6474):818.
