Abstract
Ilex × attenuata ‘Fosteri’ is an important ornamental plant widely distributed in mid-southern China and south-eastern United States. In this study, we assembled the complete chloroplast (cp) genome of I. attenuata by high-throughput sequencing and bioinformatics. The full length of cp genome was 157,833 bp with 37.63% overall GC content, which contained two inverted repeats (IR) of 26,093 bp separated by a large single-copy (LSC) and a small single copy (SSC) of 87,188 bp and 18,459 bp, respectively. The cp genome contained 135 genes, including 88 protein-coding genes, 8 rRNA genes and 39 tRNA genes. Phylogenetic tree showed that the close relationship of three species of Ilex (I. attenuata, I. viridis and I. szechwanensis) in the Aquifoliaceae family.
Ilex × attenuata ‘Fosteri’ (Foster, 1940s), also known as Foster’s Holly, is an artificial hybrid between I. cassine and I. opaca. Foster’s Holly is a small evergreen tree that is densely pyramidal silhouette. With the characteristics of dark olive-green leaves and persistent bright-red berries, it was planted as an important ornamental plant. However, due to the similar flowers and fruits with other Ilex species and cultivars, it is not easy to identify and classify by morphology. With the rapid development of the sequencing technologies, the chloroplast genome has been used to a greater extent in species identification and phylogenetic relationships in plants (Rogalski et al. Citation2015; Tonti-Filippini et al. Citation2017). Here, we reported and characterized the complete chloroplast genome of I. attenuata in an effort to provide genomic resources useful for promoting its conservation and utilization.
Fresh leaves of I. attenuata was collected from Nanjing Botanical Garden, Mem. Sun Yat-sen (E118_83, N32_06), Nanjing, China. The voucher specimen was deposited at the Institute of Botany, Jiangsu Province and Chinese Academy of Science (http://www.cnbg.net/, Hong Chen, [email protected]) under the voucher number NBGJIB-Ilex-0039. Total DNA was extracted using the GMS16011.2.1 Kit (Genmed Scientifics Inc., USA) according to manufacturer’s instructions. After the detection of DNA purity and integrity, high-quality DNA was used to library construction and sequenced using Illumina Noveseq with paired-end 150 strategy. A total of 6357.3 Mb raw data were generated, and 6149.9 Mb clean data were used for the cp genome de novo assembly by NOVOPlasty v3.3 (Dierckxsens et al. Citation2017). Finally, the cp genome annotation was performed by Geseq (Michael et al. Citation2017) combined with manual correction. The complete cp genome sequence was submitted to Genbank under accession number of MW528026.
The cp genome of I. attenuata was a typical quadripartite structure with a length of 157,833 bp, containing two inverted repeat (IR) regions of 26,093 bp, separated by a large single-copy (LSC) and a small single-copy (SSC) region of 87,188 bp and 18,459 bp, respectively. The overall GC content of the cp genome was 37.63%. A total of 135 genes were predicted, including 88 protein-coding genes, 8 rRNA genes and 39 tRNA genes. Eight protein-coding genes, four rRNA genes and seven tRNA genes are duplicated in IR regions. Besides, fifteen genes contained two exons and three genes (clpP, ycf3 and rps12) contained three exons.
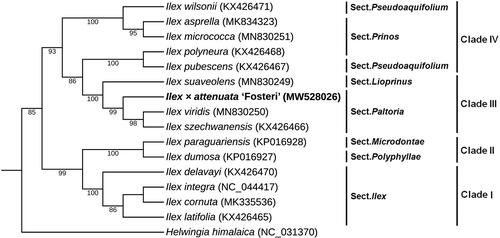
To explore the phylogenetic position and evolutionary relationship of I. attenuata, we selected 14 Ilex species and one Helwingia himalaica as outgroup for the phylogenetic analysis (Yao et al. Citation2016; Cascales et al. Citation2017; Park et al. Citation2019; Su et al. Citation2020). The phylogeny was constructed by maximum likelihood (ML) method based on 78 common protein-coding genes extracted from the complete cp genomes of 16 species using PhyML version 3.0 software (Liu et al. Citation2019). Bootstrap values were estimated from 1000 replicates. In , the phylogenetic tree revealed the presence of four clades within Ilex, in agreement with the fossil record (Yao et al. Citation2021), and also fit well with recent reports on plastid phylogenetic analysis (Yao et al. Citation2016; Su et al. Citation2020). I. attenuata was clustered with I. viridis and I. szechwanensis in section Paltoria in clade III, indicating that it has a relatively close relationship with the two Ilex species. The cp genome sequence of I. attenuata will provide a useful resource for the conservation genetics of this species as well as for building the phylogenetic relationships of Aquifoliaceae.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MW528026. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA690233, SRR13376222, and SAMN17248681, respectively.
Additional information
Funding
References
- Cascales J, Bracco M, Garberoglio MJ, Poggio L, Gottlieb AM. 2017. Integral phylogenomic approach over Ilex L. species from southern South America. Life. 7:47.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Liu J, Champer J, Langmüller AM, Liu C, Chung J, Reeves R, Luthra A, Lee YL, Vaughn AH, Clark AG, et al. 2019. Maximum likelihood estimation of fitness components in experimental evolution. Genetics. 211(3):1005–1017.
- Michael T, Lehwark P, Pellizzer T, Ulbricht-Jones E, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45:6–11.
- Park J, Kim Y, Nam S, Kwon W, Xi H. 2019. The complete chloroplast genome of horned holly, Ilex cornuta Lindl. & Paxton (Aquifoliaceae). Mitochondrial DNA Part B. 4(1):1275–1276.
- Rogalski M, Vieira L, Fraga H, Guerra M. 2015. Plastid genomics in horticultural species: importance and applications for plant population genetics, evolution, and biotechnology. Front Plant Sci. 6:586.
- Su T, Zhang M, Shan Z, Li X, Zhou B, Wu H, Han M. 2020. Comparative survey of morphological variations and plastid genome sequencing reveals phylogenetic divergence between four endemic Ilex species. Forests. 11(9):964.
- Tonti-Filippini J, Nevill P, Dixon K, Small I. 2017. What can we do with 1000 plastid genomes? Plant J. 90(4):808–818.
- Yao X, Song Y, Yang JB, Tan YH, Corlett RT. 2021. Phylogeny and biogeography of the hollies (Ilex L., Aquifoliaceae). J Syst Evol. 59(1):73–82.
- Yao X, Tan YH, Liu YY, Song Y, Yang JB, Corlett RT. 2016. Chloroplast genome structure in Ilex (Aquifoliaceae). Sci Rep. 6:28559.