Abstract
Microchironomus tabarui Sasa, 1987 is an important bioindicator for freshwater ecosystem monitoring. Although COI barcode analyes have been performed on M. tabarui, the mitogenome of this taxon has not been assembled and analyzed. Here the complete mitogenome of M. tabarui was sequenced and analyzed to confirm the systematic and phylogenetic history of this species. The mitogenome is 15,667 bp long with high A + T content and consists of 13 protein-coding genes, 22 tRNA genes, two rRNA genes, and a noncoding control region. The phylogenomic analysis support monophyletic Chironominae and close relationship between M. tabarui and Chironomus. Our results indicate that mitogenomes showed strong signals in phylogenetic reconstructions at the genus level of Chironominae.
Microchironomus tabarui Sasa, 1987 is classified in the subfamily Chironominae, Chironomidae, one of the most diverse group of aquatic Diptera with around 6300 described species (P. Ashe, personal communication). Due to their high species diversity and ability to inhabit different types of water bodies, chironomid larvae are important bioindicators for freshwater ecosystem monitoring (Ferrington Citation2008). Due to variable morphological features, entomologists face great challenges in identifying chironomids. Mitogenomic data have been broadly used in molecular identification and phylogenetic studies of Diptera (e.g., Yan et al. Citation2019; Miao et al. Citation2020; Li et al. Citation2020). However, only a few Chironomidae mitogenomes have been deciphered (Beckenbach Citation2012; Kim et al. Citation2016; Deviatiiarov et al. Citation2017; Lei et al. Citation2021; Zheng et al. Citation2021). In this study, we provide the first complete mitochondrial genome of M. tabarui.
The adult male of M. tabarui was collected from Hengshui, Hebei, China (37.651626°N, 115.650831°E) on 1 September 2020. Total genomic DNA was extracted from the muscle tissues of the head and thorax of an adult using the DNeasy Blood and Tissue kit (QIAGEN Sciences, Valencia, CA, USA). The DNA and voucher specimen of M. tabarui is deposited in the College of Life Sciences, Nankai University, Tianjin, China (https://sky.nankai.edu.cn, Xiao-Long Lin, [email protected]) under the voucher number XL3993. COI of M. tabarui (GenBank accession: KJ188136) was used as bait to iterate and assemble the mitogenome of M. tabarui. The genomic DNA was subsequently pooled with other insect species and sequenced using the Illumina Nova6000 (PE150, Illumina, San Diego, CA) platform with an insert size of 350-bp and a paired-end 150-bp sequencing strategy at Novogene Co., Ltd. (Beijing, China). Four Gb clean data were obtained from the library after trimming using Trimmomatic (Bolger et al. Citation2014). The software IDBA-1.1.1 (Peng et al. Citation2012) was employed to assemble the data with similarity set to be 0.98. The mitogenome of M. tabarui was then identified using a Blast search (Altschul et al. Citation1990) with COI as the bait sequence (Crampton-Platt et al. Citation2015), and the percentage of identical matches was 100%. The mitogenome annotation was conducted following Zheng et al. (Citation2020).
The mitogenome of M. tabarui is 15,667 bp in length (GenBank accession No. MZ261913), containing 13 protein-coding genes (PCGs), two ribosomal RNA genes, 22 transfer RNA genes, and one noncoding control region. The overall nucleotide composition was 39.8% of A, 36.6% of T, 14.2% of C, 9.4% of G, 76.4% of A + T content. Most of the 13 PCGs used ATN as the start codon (ATC for ATP8; ATG for ATP6, COII, COIII, CytB, ND4 and ND4L; ATT for ND2, ND3 and ND6; GTG for ND5; TTG for COI and ND1). The stop codon TAA is assigned to the 13 PCGs. Gene arrangement of the 13 PCGs is identical to that of other known Chironomidae mitogenomes. Nucleotide composition of M. tabarui is similar with other known Chironomidae mitogenomes, with a high A + T bias.
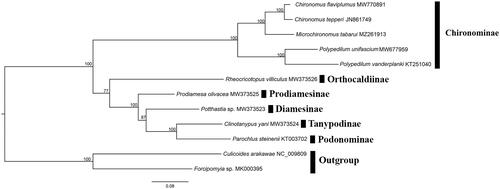
Ten mitochondrial genomes of Chironomidae and two of Ceratopogonidae available from GenBank were mined for the phylogenetic analysis. The sequences were concatenated with alignments of 13 PCGs using the default settings in MAFFT (Katoh and Standley Citation2013). The maximum-likelihood (ML) reconstruction was performed using IQ-TREE (Nguyen et al. Citation2015) with 1000 bootstraps replicates and thePMSF acid substitution model. In addition, Culicoides arakawae and Forcipomyia makanensis were designated as the outgroups. The result () clearly shows that Chironominae formed a monophyletic group. Microchironomus tabarui is sister to the genus Chironomus based on mitogenomics, which is concordant with morphology. This work provides molecular characterizations of M. tabarui and contributes to the phylogenetic analysis of Chironomidae.
Disclosure statement
No potential competing interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession No. MZ261913. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA731346, SUB9688437, SAMN19277824, respectively.
Additional information
Funding
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol. 215(3):403–410.
- Beckenbach AT. 2012. Mitochondrial genome sequences of Nematocera (lower Diptera): evidence of rearrangement following a complete genome duplication in a winter crane fly. Genome Biol Evol. 4(2):89–101.
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120.
- Crampton-Platt A, Timmermans MJ, Gimmel ML, Kutty SN, Cockerill TD, Vun Khen C, Vogler AP. 2015. Soup to tree: the phylogeny of beetles inferred by mitochondrial metagenomics of a Bornean rainforest sample. Mol Biol Evol. 32(9):2302–2316.
- Deviatiiarov R, Kikawada T, Gusev O. 2017. The complete mitochondrial genome of an anhydrobiotic midge Polypedilum vanderplanki (Chironomidae, Diptera). Mitochondrial DNA A DNA Mapp Seq Anal. 28(2):218–220.
- Ferrington LC. 2008. Global diversity of non-biting midges (Chironomidae; Insecta-Diptera) in freshwater. Hydrobiologia. 595(1):447–455.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kim S, Kim H, Shin SC. 2016. Complete mitochondrial genome of the Antarctic midge Parochlus steinenii (Diptera: Chironomidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27(5):3475–3476.
- Lei T, Song C, Zhu XD, Xu BY, Qi X. 2021. The complete mitochondrial genome of a non-biting midge Polypedilum unifascium (Tokunaga, 1938) (Diptera: Chironomidae). Mitochondrial DNA B Resour. 6(8):2212–2213.
- Li XY, Yan LP, Pape T, Gao YY, Zhang D. 2020. Evolutionary insights into bot flies (Insecta: Diptera: Oestridae) from comparative analysis of the mitochondrial genomes. Int J Biol Macromol. 149:371–380.
- Miao X, Huang J, Menzel F, Wang Q, Wei Q, Lin XL, Wu H. 2020. Five mitochondrial genomes of black fungus gnats (Sciaridae) and their phylogenetic implications. Int J Biol Macromol. 150:200–205.
- Nguyen LT, Schmidt HA, Haeseler AV, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Peng Y, Leung H, Yiu S, Chin F. 2012. IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics. 28(11):1420–1428.
- Yan L, Pape T, Elgar MA, Gao Y, Zhang D. 2019. Evolutionary history of stomach bot flies in the light of mitogenomics. Syst Entomol. 44(4):797–809.
- Zheng CG, Ye Z, Zhu XX, Zhang HG, Dong X, Chen P, Bu WJ. 2020. Integrative taxonomy uncovers hidden species diversity in the rheophilic genus Potamometra (Hemiptera: Gerridae). Zool Scr. 49(2):174–186.
- Zheng CG, Zhu XX, Yan LP, Yao Y, Bu WJ, Wang XH, Lin XL. 2021. First complete mitogenomes of Diamesinae, Orthocladiinae, Prodiamesinae, Tanypodinae (Diptera: Chironomidae) and their implication in phylogenetics. PeerJ. 9:e11294.