Abstract
In this study, we sequenced and annotated the complete mitochondrial genome of Ficedula hyperythra (Blyth, 1843). The mitogenome of F. hyperythra is 16,819 bp in length and contains 13 protein-coding, 22 transfer RNA, and 2 ribosomal RNA genes. Phylogenetic analysis based on 13 concatenated protein-coding genes showed that F. hyperythra clustered with other Ficedula species and had a close relationship with F. albicilla.
The genus Ficedula has become a model system for studies of avian speciation, genomics, biogeography, and the evolution of migratory behavior (Moyle et al. Citation2015). The snowy-browed flycatcher, Ficedula hyperythra (Blyth, 1843) is one of species in Ficedula and was reported to be not monophyletic (Moyle et al. Citation2015). The whole mitochondrial genome (mitogenome) will be useful to better understand the phylogenetic context, genetic diversity, and geographic distribution of this species. In this study, we determined a complete mitogenome of F. hyperythra using next-generation sequencing technology.
The sample was collected from Leshan, Sichuan, China (28°42′N, 103°03′E). A specimen was deposited at Southwest Minzu University (Nan Yang, [email protected]) under the voucher number SWMZU-JWY-B24101. A small amount of muscle tissue was shipped to Sangon (Shanghai, China) for genomic extraction and sequencing library construction. Whole genome sequencing was performed to generate 150 bp paired-end reads on the HiSeq 2500 platform (Illumina). After quality control, de novo assembly of clean reads was conducted using SPAdes v3.14.1 (Prjibelski et al. Citation2020). Genomic features were annotated with the MITOS web server (Bernt et al. Citation2013). The final assembled mitogenome was deposited in GenBank of NCBI with accession number MW795347.
The mitogenome of F. hyperythra was 16,819 bp in length and contained 13 protein-coding genes (PCGs), 22 transfer RNA genes, two ribosomal RNA genes and one putative control region. The nucleotide composition is significantly biased 29.27% A, 33.19% C, 14.98% G, 22.55% T, with a G + C content of 48.18%. The gene arrangement and composition were identical with the mitogenome of F. albicilla (Zhang and Lu Citation2019), exhibiting a typical vertebrate mitochondrial gene arrangement. The start codon for all PCGs was ATG, except COX1, which had ATC as the start codon. We identified three different complete termination codons: AGG for COX1, TAG for ND1 and ND6, TAA for ND2, COX2, ATP8, ATP6, ND3, ND4L, ND5, and Cytb. For COX3 and ND4, the termination codon was incomplete (T–).
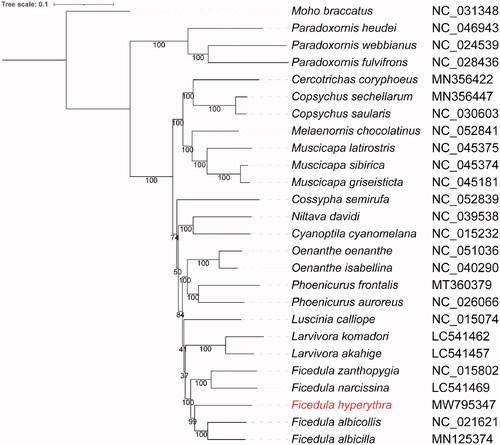
Phylogenetic analysis for F. hyperythra and the other 24 Muscicapidae species was conducted by concatenating nucleotide sequences of 13 PCGs. One species of Mohoidae (Moho braccatus) was used as an outgroup. Multiple sequence alignment was performed using MAFFT v7.475 (Katoh and Standley Citation2013). A maximum-likelihood phylogenetic tree was constructed using IQtree v1.6.12 (Nguyen et al. Citation2015) with 1000 bootstrap replicates and TVM + F+R4 substitution model suggested by modelFinder (Kalyaanamoorthy et al. Citation2017). The ML phylogenetic tree illustrated that our sample clustered with other Ficedula species and had a close relationship with F. albicilla ().
Disclosure statement
No potential competing interest was reported by the authors.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession MW795347. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA753463, SRR15403527, and SAMN20695163 respectively.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Kalyaanamoorthy S, Minh BQ, Wong TKF, Von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Moyle RG, Hosner PA, Jones AW, Outlaw DC. 2015. Phylogeny and biogeography of Ficedula flycatchers (Aves: Muscicapidae): novel results from fresh source material. Mol Phylogenet Evol. 82 Pt A(Pt A):87–94.
- Nguyen L-T, Schmidt HA, Von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Prjibelski A, Antipov D, Meleshko D, Lapidus A, Korobeynikov A. 2020. Using SPAdes De Novo Assembler. Curr Protoc Bioinformatics. 70(1):e102.
- Zhang XR, Lu CH. 2019. The complete mitochondrial genome of red-throated flycatcher Ficedula albicilla (Passeriformes: Ficedula). Mitochondrial DNA B Resour. 4(2):3322–3323.