Abstract
Rice flower carp (Cyprinus carpio rubrofuscus) is a subspecies of Cyprinus carpio distributed in South China. In this study, we sequenced the mitochondrial genome of C. carpio rubrofuscus using Illumina HiSeq. The complete mitochondrial genome of C. carpio rubrofuscus was 16,582 bp in length, including 13 protein-coding genes, 22 transfer RNA genes, and two ribosomal RNA genes. The contents of the four bases in the mitochondrial DNA were A (31.89%), T (24.82%), C (27.53%), and G (15.76%). Phylogenetic analysis showed that C. carpio rubrofuscus was clustered with Cyprinus carpio and its varieties.
Rice flower carp (Cyprinus carpio rubrofuscus) is a subspecies of Cyprinus carpio found widely in South China, such as the Pearl River and Hainan Island regions, where it inhabits the grassy or muddy edges of rivers (Wu Citation1977). The species is also commonly cultured with rice crops in western and northern Guangdong, China (Wu Citation1977). The body of C. carpio rubrofuscus varies from striped and flat to round and thick and its color varies from steel and light gray to golden red. The species is not large (∼50–250 g) and is characterized by cold-resistance, alkali-resistance, and hypoxic tolerance (Ma et al. Citation2018). However, research on this species remains limited. Ma et al. (Citation2018, Citation2019) studied the morphological characteristics and genetic diversity of C. carpio rubrofuscus. The complete mitochondrial genome (mitogenome) of C. carpio rubrofuscus is an important resource for evolutionary research. In this study, we report on the complete mitogenome of C. carpio rubrofuscus and analyze its phylogenetic relationships within Cyprinus.
Specimens (voucher no. QP20190316-9) were collected from the Beijiang River (23°19′ N, 113°16′ E), Qingyuan, Guangdong, China, and were stored in the herbarium of Qingyuan Polytechnic (Guangdong, China). Muscle samples of C. carpio rubrofuscus were dissected and preserved at −80 °C until use. The muscle tissue was used for genomic DNA extraction with an E.Z.N.A.® Tissue DNA Kit (Omega Bio-Tek, Guangzhou, China) following the manufacturer’s specifications. The mtDNA was sequenced using Illumina HiSeq (Illumina Inc., San Diego, CA). Clean data were acquired and assembled using SPAdes v3.15.2 (Bankevich et al. Citation2012) and PRICE (Ruby et al. Citation2013). MITO (http://mitos.bioinf.uni-leipzig.de/index.py) (Bernt et al. Citation2013) and ORF finder (https://www.ncbi.nlm.nih.gov/orffinder/) were used to identify and annotate protein-coding, transfer RNA (tRNA), and ribosomal RNA (rRNA) genes. Phylogenetic analysis was conducted using maximum-likelihood (ML) in MEGA X (Kumar et al. Citation2018).
The mitogenome of C. carpio rubrofuscus was 16,582 bp in length (GenBank accession number: MW969691) and contained 13 protein-coding, 22 tRNA, and two rRNA genes. Of the 37 genes, 28 were encoded by the heavy strand and nine, including one protein-coding (ND6) gene and eight tRNA genes, were encoded by the light strand. The A, G, C, and T contents of the heavy strand were 31.89%, 15.76%, 27.53%, and 24.82%, respectively, with a high C + G content of 43.29%. Most protein-coding genes had an ATG start codon, except Cox1 (initiated with GTG). Seven protein-coding genes (ND1, Cox1, ATP6, Cox3, ND4L, ND5, and ND6) contained a TAA stop codon, three protein-coding genes (ND2, ND3, and ATP8) contained a TAG stop codon, and three protein-coding genes (Cytb, ND4, and Cox2) contained an incomplete T– stop codon. 16S rRNA and 12S rRNA were 1679 bp (56.40% AT content) and 953 bp (51.42% AT content) in length, respectively. All tRNA genes ranged from 67 to 76 bp in size.
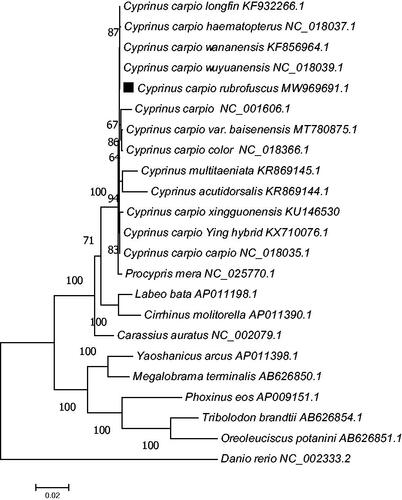
A phylogenetic tree was constructed to validate the phylogenetic position of C. carpio rubrofuscus. The complete concatenated protein sequence which was coded by 13 protein-coding genes of another 22 Cyprinidae varieties was downloaded from GenBank. A phylogenetic tree was constructed using the ML method and Jones–Taylor–Thornton (JTT) matrix-based model, with a bootstrap of 1000 replicates (). The phylogenetic tree showed that all Cyprinus carpio individuals and subspecies were clustered together. In conclusion, our study described the complete mitogenome of C. carpio rubrofuscus and analyzed its phylogenetic position within the Cyprinus genus. This research should contribute to further investigations on the molecular evolution and conservation of this species.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
Mitogenome data supporting this study are openly available in GenBank at: https://www.ncbi.nlm.nih.gov/nuccore/MW969691.1/. Associated BioProject, SRA, and BioSample accession numbers are http://www.ncbi.nlm.nih.gov/bioproject/742061, https://www.ncbi.nlm.nih.gov/sra/PRJNA742061, and https://www.ncbi.nlm.nih.gov/biosample/SAMN19923788, respectively.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Ma DM, Huang ZH, Zhu HP, Xie J. 2019. Morphological characteristics and genetic analysis of the rice flower carp in the northern region of Guangdong Province. Prog Fish Sci. 40(2):33–42.
- Ma DM, Su HH, Zhu HP, Huang ZH. 2018. Genetic diversity and genetic structure analysis of different selective breeding generations in Cyprinus carpio rubrofuscus using microsatellite markers. Acta Hydrobiol Sin. 42(5):887–895.
- Ruby JG, Bellare P, Derisi JL. 2013. PRICE: software for the targeted assembly of components of (Meta) genomic sequence data. G3. 3(5):865–880.
- Wu XW. 1977. The fish of Cyprinidae in China. Shanghai: Shanghai People's Publishing House; p. 408–428.