Abstract
Euchresta tubulosa Dunn not only is a national second-level protected wild plant in China, but also has a long history as a source plant in traditional Chinese medicine. The chloroplast (cp) genome of E. tubulosa was 154,102 bp, consisting of a large single-copy region (LSC: 92,877 bp), a small single-copy region (SSC: 36,645 bp), and a pair of inverted repeat regions (IRb and Ira: 12,290 bp, respectively). These sequences encoded 123 genes, including 78 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. The phylogenetic analysis showed that E. tubulosa is close to Lupinus species.
Euchresta tubulosa Dunn, with dried roots known as ‘Shan Dou Gen’ in traditional Chinese medicine, is an evergreen vine-like shrub of the genus Euchresta in Fabaceae. Euchresta tubulosa is a national second-level protected wild plant in China, which is scattered in parts of Guangdong, Guangxi, Sichuan and Zhejiang of China, as well as in Japan. It not only has the effects of clearing away heat, detoxifying, reducing swelling and pain, but also has the effects of treating enteritis, diarrhea, abdominal pain, and stomach pain. Euchresta tubulosa is one of the common Chinese herbal medicines in the folk (Lin et al. Citation2009; Li et al. Citation2011). Modern pharmacological studies have found that E. tubulosa contains flavonoids (Matsuura et al. Citation1993), alkaloids (Li et al., Citation2019), steroids and other compounds (Li et al. Citation2014). At the same time, Euchresta species has the activities of anti-tumor, inducing cell apoptosis, central inhibition, antioxidant (Toda and Shirataki Citation2006), regulating blood lipids, antibacterial and other pharmacological activities (Li and Li Citation2016). However, as an important medicinal plant, E. tubulosa has not been reported on genome related information. In this study, we report the first complete chloroplast (cp) genome sequence of E. tubulosa, which not only helps to better understand its evolution and population genetics, but also provides important information for the phylogeny of Euchresta.
Fresh leaves of E. tubulosa were collected from Nanchuan, Chongqing, China (107°21′E, 29°14′N, 600 m). The voucher specimen was conserved in Chongqing Institute of Medicinal Plant Cultivation (accession number: CIMPC-RFM-20210403, Contact person: Fengming Ren; Email: [email protected]). In the experiment, the high-quality whole genomic DNA was extracted by the kit method (Beijing Kinco Biological Company). The purity and integrity of the DNA were analyzed by Nanodrop (Thermo Fisher Scientific) and agarose gel electrophoresis. Total DNA was used to generate libraries with insert size of 350 bp and was generated raw pair-end reads by Illumina Hiseq 2500 Platform (Illumina, Hayward, CA, USA).
Low-quality readings and adapters in the raw data are deleted by trimmomati (version 0.35) with default paprameters (Bolger et al. Citation2014). About 15 G clean data was obtained. Using the clean data with 150 bp paired-end read lengths obtained from the raw data, a cp genome was assembled by NOVOPlasty (version 4.1) with the default parameters (Nicolas et al. Citation2016) and annotated by CPGAVAS2 (http://47.96.249.172:16019/analyzer/home) (Shi et al. Citation2019). Finally, manually correct the start and stop codons of protein-coding genes.
The cp sequence of E. tubulosa was submitted to NCBI, and the accession number was MZ323108. The cp genome of E. tubulosa is 154,102 bp in size, containing four regions: large single-copy region (LSC: 92,877 bp), small single-copy region (SSC: 36,645 bp), and a pair of inverted repeat regions (IR: 12,290 bp). The GC contents of whole genome and each genomic regions were 36.27% (whole genome), 34.18% (LSC region), 40.12% (SSC region), 38.41% (IR region). A total of 123 genes were annotated, including 78 protein-coding genes, 37 tRNA genes, and 8 rRNA genes.
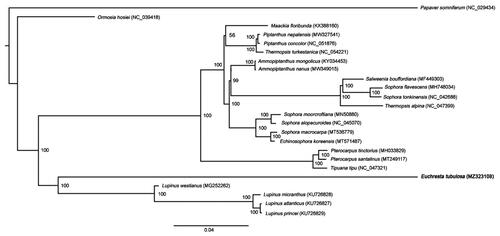
To investigate its taxonomic status, additional 22 species of Fabaceae from the NCBI website were selected to study. Papaver somniferum was used as the outgroup. The reference sequences were compared by MAFFT software with the parameter of ‘–auto’ (Katoh and Standley Citation2013). Based on the aligned sequences, a phylogenetic tree was built with 1000 bootstrap replicates by IQ-TREE (version 1.6.12) (Lam-Tung et al. Citation2015). As shown in the phylogenetic tree (), all species were clustered on the correct clades. Phylogenetic analysis showed that E. tubulosa clustered in a separated branch in the Fabaceae, which was the sister of Lupinus species. The new provided molecular data is helpful to illuminate the Fabaceae evolution.
Acknowledgements
We thank Prof. Qiuping Tan for providing plant material.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, under the accession number MZ323108. The associated Bio-Project, Bio-Sample and SRA numbers are PRJNA543381, SAMN19414659 and SRR14682996 respectively.
Additional information
Funding
References
- Bolger AM, Marc L, Bjoern U. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability . Mol Biol Evol. 30(4):772–780.
- Lam-Tung N, Schmidt HA, Arndt VH, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Li HC, Yuan DP, Yuan CY. 2011. Research progress on endangered medicine Euchresta tubulosa of Tujia Nationality. J Hubei Univ Nationalities. 28(04):71–73.
- Li HD, Yuan DP, Liu Y. 2014. Research progress on chemical constituents in plants of Euchresta J. Benn and their biological activities. Chin Traditional Herbal Drugs. 45(23):3486–3493.
- Li QS, Li YB. 2016. Purification and extraction of Euchresta Japonica glycoside and aglucon. World Latest Med Information. 86:85–86.
- Li WX, Wang H, Dong AW. 2019. Preparative separation of alkaloids from stem of Euchresta tubulosa Dunn. by high-speed counter-current chromatography using stepwise elution. Molecules. 24(24):4602.
- Lin MX, Han F, Liu ZY, Zhang J, Shen J, Ren MB. 2009. Research on the folk anticancer drug Euchresta japonica. Modern Chin Med Res Practice. 23(05):26–28.
- Matsuura N, Iinuma M, Tanaka T, Mizuno M. 1993. Flavonoids and a benzofuran in roots of Euchresta tubulosa. Phytochemistry. 33(3):701–705.
- Nicolas D, Patrick M, Guillaume S. 2016. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):4.
- Shi L, Chen H, Jiang M, Wang L, Wu X, Huang L, Liu C. 2019. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47(1):65–73.
- Toda S, Shirataki Y. 2006. Inhibitory effect of prenylated flavonoid in Euchresta japonica. and Artocarpus heterophyllus on lipid peroxidation by interaction of hemoglobin and hydrogen peroxide. Pharml Biol. 44(4):271–273.