Abstract
Diospyros kaki cv. Luotiantianshi is a rare germplasm of Diospyros Linn in the world. In this study, we generated the complete chloroplast (cp) genome of D. kaki cv. Luotiantianshi. The complete cp genome was 157,773 bp in length, containing a large single copy region (LSC) of 87,066 bp, a small single copy region (SSC) of 18,529 bp, and two inverted repeat (IR) regions of 26,089 bp. The new sequence has a total of 131 genes, including 86 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. Further, phylogenetic analysis showed that the D. kaki cv. Luotiantianshi has a close relationship with Diospyros kaki. This study provides important information for future evolution, genetic and molecular biology studies of Diospyros.
Diospyros kaki Thunb 1780 cv. Luotiantianshi, also is called Chinese PCNA (pollination-constant non-astringent) type persimmon (Diospyros kaki Thunb 1780), only distributes in Luotian and Macheng of Hubei province. Diospyros kaki cv. Luotiantianshi, which is morphologically and genetically different from Japanese pollination constant and non-astringent (JPCNA) or other persimmons by natural de-astringency model of fruits (Yonemori et al. Citation2005), is a rare germplasm of Diospyros Linn in the world (Zhang et al. Citation2016). In the present study, the complete chloroplast genome of D. kaki cv. Luotiantianshi was obtained using the next-generation sequencing (NGS) technologies and a phylogenetic analysis of this species and its relatives was carried out.
Total DNA was extracted from 5 g fresh leaves gathered from Pinghu town in Luotian county, Huanggang city, Hubei province of China (115°22'2″E, 30°54'49″N) using CTAB method (Doyle and Doyle Citation1987). A specimen was deposited at Research Institute of Subtropical Forestry, Chinese Academy of Forestry (URL: http://risfcaf.caf.ac.cn/, contact person: Bang-chu Gong, email: [email protected]) under the voucher number: MBLS-LTTS84. The DNA libraries was constructed and sequenced on Illumina HiseqXten platform (Illumina, San Diego, CA) for 150 bp paired-end sequencing by Biodata Biotechnologies Inc. (Hefei, China). The cp genome was assembled using a de novo strategy according to the work of Xu et al. (Citation2020).
The size of the complete chloroplast genome of D. kaki cv. Luotiantianshi was 157,773 bp, which was smaller than D. kaki (157,784 bp) and larger than Diospyros oleifera Cheng 1935 (157,724 bp) (Fu et al. Citation2016). The cp genome of D. kaki cv. Luotiantianshi was composed of a large single-copy region (LSC, 87,066 bp), a small single-copy region (SSC, 18,529 bp), and two inverted repeats (IR, 26,089 bp). The total GC content of new chloroplast genome was 37.4%, and while it was 43.1% of IRs, which was higher than LSC and SSC regions (35.40% and 30.80%, respectively). The new sequence has 86 protein-coding genes, was one less than that in D. kaki (87), 37 tRNA genes, and 8 rRNA genes.
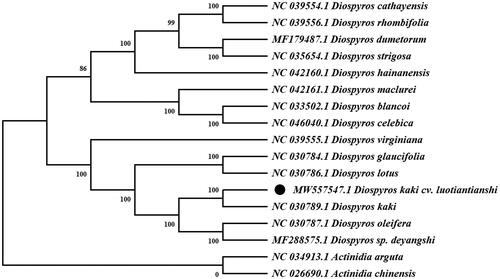
A phylogenetic analysis was carried out using whole cp genome sequences of D. kaki cv. Luotiantianshi and other 14 species in Diospyros. Meanwhile, two species from the Actinidia (Actinidia arguta Sieb. & Zucc 1864 and Actinidia chinensis Planch 1847) were used as outgroups. The sequences were aligned using MAFFT version 7 (Kazutaka and Standley Citation2013). The maximum-likelihood (ML) method base on GTR + G + I model was used to construct the phylogenetic tree with Mega7 using 1000 bootstrap (Kumar et al. Citation2016). The results () indicated that D. kaki cv. Luotiantianshi has a close relationship with D. kaki under 100% bootstrap support values. The complete cp genome sequence of D. kaki cv. Luotiantianshi can be great benefit to further study on its population genetics and molecular breeding.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov under the accession no. MW557547.1 The raw sequence data used in this research were deposited successfully with registered numbers of associated BioProject, SRA, and Bio-Sample:PRJNA701835, SRX10097592, and SAMN17914800, respectively
Additional information
Funding
References
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Kazutaka K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 3(4):772–780.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Fu J, Liu H, Hu J, Liang Y, Liang J, Wuyun T, Tan F. 2016. Five complete chloroplast genome sequences from Diospyros: genome organization and comparative analysis. PLOS One. 11(7):e0159566.
- Xu Y, Gong B, Wu K. 2020. Characterization of the complete chloroplast genome sequence of Pachira macrocarpa Walp. (Malvales). Mitochondrial DNA Part B. 5(2):1198–1199.
- Yonemori K, Ikegami A, Kitajima A, Luo Z, Kanzaki S, Sato A, Yamada M, Yang Y, Wang R. 2005. Existence of several pollination constant non-astringent type persimmons in China. Acta Hortic. 685(685):77–84.
- Zhang N, Xu J, Mo R, Zhang Q, Luo Z. 2016. Androecious genotype ‘Male 8’ carries the CPCNA gene locus controlling natural deastringency of Chinese PCNA persimmons. Hortic Plant J. 2(6):309–314.