Abstract
The mitochondrial genome (mitogenome) of Gallinago gallinago gallinago Linnaeus, 1758 was determined by the high-throughput data. The assembled mitogenome was 16,919 bp in length, with a 58.7% A + T content and GC skew of −0.3850. Among 13 PCGs, an unusual start codon (GTG) was identified for the COX1 gene, and incomplete stop codons (T-) were found in the COX3, ND2 and ND4 genes. The function of a cytosine insertion at site 174 in the ND3 gene and its phylogenetic significance are worthy of further scrutiny. In the control region (CR), thirteen 15-bp simple sequence repeats were found in G. g. gallinago. Phylogenetic analysis indicated that Gallinago was clustered at the basal position of the Scolopax clade and that the monophyly of Gallinago was also recovered. The mitogenome data of G. g. gallinago provides useful resources for further studying the evolution of Scolopacidae.
The common snipe, Gallinago gallinago, is a medium-sized, ground-nesting shorebird that feeds by probing in mud or soil to locate invertebrates with its long bill. This species occurs on all continents except Antarctica and Australia, but populations have been declining in Europe because of the drainage of bogs and marshy grassland (Cramp and Simmons Citation1983). Most species of birds separate molting from other energy-demanding activities, such as migration or reproduction. G. gallinago is an exception, as during the first autumn migration, many young snipe initiate their post-juvenile molt, which includes the replacement of body feathers, lesser and median wing coverts, tertials, and rectrices (Podlaszczuk et al. Citation2017). G. gallinago is a grassland waterbird characteristic of agricultural meadows and a member of one of the most threatened bird guilds (Regos et al. Citation2020). As an indicator species of the ecological environment, research on G. gallinago has mostly focused on macroscopic ecology (Green Citation1988; Henderson et al. Citation2002; Włodarczyk et al. Citation2018). Limited molecular data hamper phylogenetic and evolutionary studies in G. gallinago. Here, the complete mitogenome of G. g. gallinago has been reported with general features. These results are used to explore the potential for improving breeding success by habitat management.
G. g. gallinago in this study was a female adult that died naturally during the breeding season from Lantian County, Xi’an (34°20′7ʺN, 109°22′54ʺE). The specimen (voucher number: SWSZ01) was collected and identified by C. Yang, and deposited in the animal specimen museum of Shaanxi Institute of Zoology, Xi’an, Shaanxi Province, China (contacts: Chao Yang, [email protected]).
DNA was extracted by Qiagen columns (DNeasy® Blood & Tissue kit; Qiagen, Hilden, Germany) and prepared with a paired-end (2 × 150 bp) library strategy followed by next-generation sequencing (NGS) on the Illumina HiSeq Xten platform (Illumina, San Diego, CA). A total of 9,219,657 paired-end raw reads were produced. After removing regions with a Phred score of <10, 9,111,941 clean reads were obtained after quality and ambiguity controls. The clean data were assembled using MITOBim v1.9 (Hahn et al. Citation2013) with the mitogenome of G. stenura (GenBank accession no. KY056596) as a reference. Assembly of the clean reads and gene annotation were performed by Geneious 10.1.3 (Kearse et al. Citation2012) and tRNAscan-SE 2.0 (Lowe and Chan Citation2016), with a total of 210,657 mitochondrial reads mapped to the reference mitogenome, giving an average coverage of 1820.1×.
The assembled G. g. gallinago mitogenome (GenBank accession no. MZ157405) was a 16,919 bp long circular DNA with overall nucleotide frequencies of A = 32.5%, T = 26.2%, C = 28.6%, and G = 12.7% and an A + T content of 58.7%. The GC skew was −0.3850, which showed a remarkable C skew and was similar to the mitogenomes of other Charadriiformes species (Yu et al. Citation2014; Yang et al. Citation2016). With the exception of COX1, which started with GTG, all PCGs had typical ATN start codons, and all PCGs ended with a complete triplet codon (TAA, AGG, AGA, or TAG), except for COX3, ND2, and ND4, which ended with an incomplete T. All transfer RNA (tRNA) genes had typical cloverleaf secondary structures, with the exception of tRNA-Ser (AGY), in which the dihydrouridine arm formed a simple loop. The length of 12S ribosomal RNA (rRNA) was 972 bp, and that of 16S rRNA was 1598 bp, both were located between tRNA-Phe and tRNA-Leu (UUR) and separated by tRNA-Val. The CR was 1364 bp long and was located between tRNA-Glu and tRNA-Phe.
The newly sequenced species with 99.3% sequence similarity to G. gallinago (GB: MW865755), and the nucleotide differences between of them occurred mainly in the CR: length of poly-C block in domain I and number of repeat units in domain III (). In the CR, 13 (positions: 16,691–16,885) and 8 (positions: 16,680–16,799) simple sequence repeats of 5′-AAACAAACAATCAAC-3′ existed in G. g. gallinago (GB: MZ157405) and G. gallinago (GB: MW865755), respectively (software: tandem repeats finder (TRF) v4.09; Benson Citation1999). This is the main reason for the different lengths of the two sequences. More molecular sequence data generated may be applied to elucidate the population genetics of this bird species.
Figure 1. (A) Main nucleotide trait differences between G. gallinago (GenBank accession no. MW865755, MZ157405). (B) Maximum-likelihood tree obtained using RAxML v7.0.3 with 1000 nonparametric bootstrap replicates. GenBank accession numbers are indicated following species names. Numbers on the nodes are bootstrap values. ‘*’ represents no cytosine insertion.
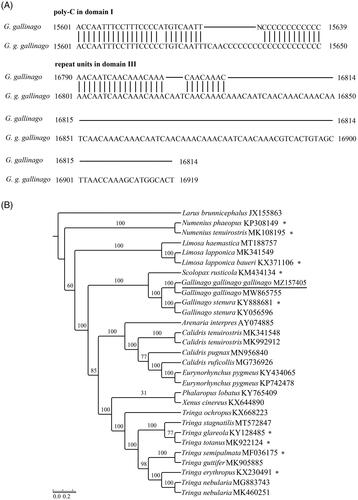
The length of the ND3 gene was 351 bp, which was similar to most of the known Scolopacidae mitogenomes in GenBank. One cytosine insertion at site 174 was revealed in the ND3 gene (nucleotide position: 9703). Classic inference suggests that this extra nucleotide may be removed by RNA editing during translation, and the function of the ND3 gene may be recovered, effectively avoiding the premature stoppage of transcription due to a frameshift mutation (Mindell et al. Citation1998). The latest hypotheses expounded that relaxed selection pressure may have allowed frameshift insertions to be tolerated for hundreds of millions of years, possibly a result of the rapid adaptive radiation of birds due to programmed translational frameshifting (Rosengarten et al. Citation2008; Russell and Beckenbach Citation2008; Haen et al. Citation2014). Species without cytosine insertion in the ND3 gene of Scolopacidae have been labeled with ‘*’ (). At the order level, species with cytosine insertion in the ND3 gene (Struthioniformes) were clustered into one branch and species without cytosine insertion in the ND3 gene clustered into another branch (Passeriformes). Species with cytosine insertion in the ND3 gene were so antiquated that they were divided out earlier. Species with/without cytosine insertion in the ND3 gene (Tinamiformes, Falconiformes, Charadriiformes, Sphenisciformes, Ciconiiformes, Galliformes, Anseriformes, etc.) clustered at the neutral position of the avian mitogenomic tree (Slack et al. Citation2007; Tamashiro et al. Citation2019). But at the family level, it seems that the clustering of related species are not hindered by cytosine insertion in the ND3 gene (). The mechanism of the extra 'C' in the ND3 gene and its phylogenetic significance on avian adaptive radiation need to be studied further.
To validate the phylogenetic position of G. g. gallinago, maximum-likelihood (ML) methods were employed to construct phylogenetic tree using RAxML v7.0.3 (Stamatakis Citation2006) based on 13 PCGs of 29 mitogenomes. The 13 PCGs sequences were aligned using Clustal v2.1 after manually removed the stop codons, and then concatenated into a combined dataset using SequenceMatrix v1.7.8. The best partitioning scheme and optimal model (models GTR + I + G and GTR + G) were analyzed in Partitionfinder v1.1.1 (Lanfear et al. Citation2012), and the robustness of the phylogenetic result was tested through bootstrap analysis with 1000 replicates (Yang et al. Citation2017). Larus brunnicephalus (GenBank accession no. JX155863) was selected as an outgroup. The topological structure showed that the monophyly of Gallinago was recovered, with the phylogeny ((G. gallinago (GB: MW865755), G. g. gallinago (GB: MZ157405)) (G. stenura (GB: KY888681), G. stenura (GB: KY056596))) () (Gibson and Baker Citation2012). We also confirmed that Gallinago was clustered at the basal position of the Scolopax clade (), which was consistent with the results of previous studies (Hu et al. Citation2017). This mitogenome would provide an important resource for further exploring the taxonomic status of Scolopacidae species.
Disclosure statement
No potential conflicts of interest were reported by the authors.
Data availability statement
The genome sequence data that support the findings of this study are openly available in NCBI GenBank (https://www.ncbi.nlm.nih.gov/) under accession no. MZ157405. The associated BioProject, BioSample, and SRA numbers are PRJNA727369, SAMN19016979, and SRR14424283, respectively.
Additional information
Funding
References
- Benson G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27(2):573–580.
- Cramp S, Simmons KEL. 1983. The birds of the Western Palearctic. Oxford: Oxford University Press.
- Gibson R, Baker A. 2012. Multiple gene sequences resolve phylogenetic relationships in the shorebird suborder Scolopaci (Aves: Charadriiformes). Mol Phylogenet Evol. 64(1):66–72.
- Green RE. 1988. Effects of environmental factors on the timing and success of breeding of common Snipe Gallinago gallinago (Aves: scolopacidae). J Appl Ecol. 25(1):79–93.
- Haen KM, Pett W, Lavrov DV. 2014. Eight new mtDNA sequences of glass sponges reveal an extensive usage of +1 frameshifting in mitochondrial translation. Gene. 535(2):336–344.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads – a baiting and iterative mapping approach. Nucl Acids Res. 41(13):e129–e129.
- Henderson IG, Wilson AM, Steele D, Vickery JA. 2002. Population estimates, trends and habitat associations of breeding Lapwing Vanellus vanellus, Curlew Numenius arquata and Snipe Gallinago gallinago in Northern Ireland in 1999. Bird Study. 49(1):17–25.
- Hu CC, Zhang CL, Sun L, Zhang Y, Xie WL, Zhang BW, Chang Q. 2017. The mitochondrial genome of pin-tailed snipe Gallinago stenura, and its implications for the phylogeny of Charadriiformes. PLOS One. 12(4):e0175244.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Lanfear R, Calcott B, Ho SY, Guindon S. 2012. Partitionfinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol Biol Evol. 29(6):1695–1701.
- Lowe TM, Chan PP. 2016. tRNAscan-SE on-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44(W1):W54–W57.
- Mindell DP, Sorenson MD, Dimcheff DE. 1998. An extra nucleotide is not translated in mitochondrial ND3 of some birds and turtles. Mol Biol Evol. 15(11):1568–1571.
- Podlaszczuk P, Włodarczyk R, Janiszewski T, Kaczmarek K, Minias P. 2017. When moult overlaps migration: moult-related changes in plasma biochemistry of migrating common snipe. Peer J. 5:e3057.
- Regos A, Vidal M, Lorenzo M, Domínguez J. 2020. Integrating intraseasonal grassland dynamics in cross-scale distribution modeling to support waterbird recovery plans. Conserv Biol. 34(2):494–504.
- Rosengarten RD, Sperling EA, Moreno MA, Leys SP, Dellaporta SL. 2008. The mitochondrial genome of the hexactinellid sponge Aphrocallistes vastus: evidence for programmed translational frameshifting. BMC Genomics. 9:33.
- Russell RD, Beckenbach AT. 2008. Recoding of translation in turtle mitochondrial genomes: programmed frameshift mutations and evidence of a modified genetic code. J Mol Evol. 67(6):682–695.
- Slack KE, Delsuc F, Mclenachan PA, Arnason U, Penny D. 2007. Resolving the root of the avian mitogenomic tree by breaking up long branches. Mol Phylogenet Evol. 42(1):1–13.
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22(21):2688–2690.
- Tamashiro RA, White ND, Braun MJ, Faircloth BC, Braun EL, Kimball RT. 2019. What are the roles of taxon sampling and model fit in tests of cyto-nuclear discordance using avian mitogenomic data? Mol Phylogenet Evol. 130:132–142.
- Włodarczyk R, Podlaszczuk P, Kaczmarek K, Janiszewski T, Minias P. 2018. Leukocyte profiles indicate nutritional, but not moulting stress in a migratory shorebird, the Common Snipe (Gallinago gallinago). J Ornithol. 159(2):345–354.
- Yang C, Wang QX, Huang Y, Xiao H. 2016. Complete mitochondrial genome of Relict Gull, Larus relictus (Charadriiformes: Laridae). Mitochondrial DNA Part A. 27(1):411–412.
- Yang C, Wang QX, Li XJ, Xiao H, Huang Y. 2017. Characterization of the mitogenomes for two sympatric breeding species in Recurvirostridae (Charadriiformes) and their phylogenetic implications. Mitochondrial DNA Part B. 2(1):182–184.
- Yu TT, Hu CC, Chang Q, Yang RY, Dong WW, Xie WL. 2014. The complete mitochondrial genome of Scolopax rusticola (Charadriiformes: Scolopacidae). Mitochondrial DNA Part A. 27(3):1800–1801.
