Abstract
Beesia deltophylla is an endemic and rare species only distributed in Xizang, China. The chloroplast genome of B. deltophylla is 157,397 bp in length, with 112 encoded genes including 78 protein-coding genes, 30 tRNA genes and 4 rRNA genes. Phylogenetic reconstruction has confirmed the placement of B. deltophylla as sister to B. calthifolia. These two species formed a clade closely to a Japan endemic species Anemonopsis macrophylla.
Beesia deltophylla C. Y. Wu is a buttercup species endemic in Xizang, China (Xiao Citation1979). It has a sister species, B.calthifolia, mainly distributed in Southwest China. Previous studies have confirmed this genus as a monophyletic group related to the Anemonopsis in Cimicifugeae (Yang et al. Citation1995, Wang et al. Citation1998, Gao et al. Citation2008). The genus Beesia is also a traditional Chinese medicine resource. The population size of B. deltophylla is extremely small and was situated in an extremely small population in the Red List of endangered species (Vié et al. Citation2009). At present, the chloroplast genome of B. calthifolia has been published (Zhai et al. Citation2019). Here, we report the complete chloroplast genome of B. deltophylla and reconstruct phylogenetic relationship to provide more molecular sequences for further research.
The samples of B. deltophylla were collected from Xizang China (29°22′ N, 95°8′ E). A specimen was deposited at Wetland College of Southwest Forestry University (http://plateauwetland.swfu.edu.cn/, Liang-Liang Yue, email: [email protected]) under the voucher number yue20190135. A sequencing library was sequenced using Illumina nova-seq 6000 platform. NGS QC ToolKit (Patel and Jain Citation2012) was utilized to filter all raw readings and obtain high-quality clean reads. The complete chloroplast genome was assembled using NOVOPlasty (Dierckxsens et al. Citation2017). The chloroplast genome of B. calthifolia (GenBank accession number: NC_041531) was used as a reference sequence for assembling. After annotating the genome using GeSeq (Tillich et al. Citation2017), GB2squine (Lehwark and Greiner Citation2019) was utilized to convert the generated chloroplast genome annotation files into feature table files and submitted to GenBank via online BankIt.
The chloroplast genome of B. deltophylla has a full length of 157,397 bp. It is generally presented as a typical quartile structure, consisting of a large single-copy region (LSC, 86,863 bp), a small single-copy region (SSC, 17,481 bp), and a pair of inverted repeat regions (IRa/b, 26,525 bp). The chloroplast genome contains 112 genes, including 78 protein-coding genes, 30 tRNA genes, and 4 rRNA genes. The GC content of the B. deltophylla chloroplast genome was 38% and the corresponding values in LSC, SSC, and IR regions were 36.6%, 32.6%, 43.2%, respectively. Annotated chloroplast genome sequence was submitted to GenBank with an accession number MZ350960.
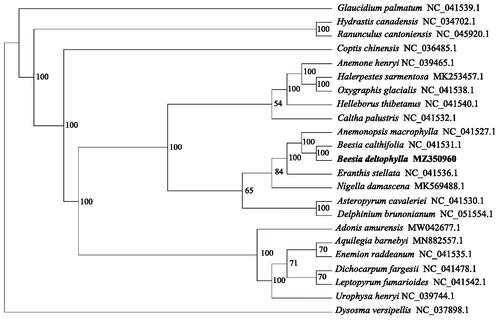
The phylogenetic analysis was based on B. deltophylla and other 22 complete chloroplast genomes published on NCBI, using the Dysosma versipellis as the outgroup. All sequences were aligned by MAFFT v7 (Katoh and Standley Citation2013) and manually adjusted in MEGA 7.0 (Kumar et al. Citation2016). The most suitable nucleotide substitution model is determined by ModelFinder (Kalyaanamoorthy et al. Citation2017), and the best model is determined as GTR + F+R4. Then use IQ-TREE 1.62 (Nguyen et al. Citation2015) to perform maximum likelihood (ML) analysis based on the best model and 5000 ultra-fast bootloaders . The ML tree showed that Beesia is a monophyletic group with a 100% bootstrap value (). Our results provide basic information for further phylogenetic and biogeographic researches on the Beesia and the Cimicifugeae.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genomic sequence data supporting the results of this study can be obtained in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov) under the accession number MZ350960. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA744067, SRR15048188, and SAMN20079128, respectively.
Additional information
Funding
References
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Gao JC, Peng Y, Yang MS, Xiao PG. 2008. A preliminary pharmacophylogenetic study of tribe Cimicifugeae (Ranunculaceae). J Syst Evol. 46(4):516–536.
- Kalyaanamoorthy S, Minh BQ, Wong TKF, Von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Lehwark P, Greiner S. 2019. GB2sequin – a file converter preparing custom GenBank files for database submission. Genomics. 111(4):759–761.
- Nguyen LT, Schmidt HA, Von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Patel RK, Jain M. 2012. NGS QC toolkit: a toolkit for quality control of next generation sequencing data. PLOS One. 7(2):e30619.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Vié JC, Hilton-Taylor C, Stuart SN. 2009. Wildlife in a changing world: an analysis of the 2008 IUCN Red List of threatened species. IUCN.
- Wang XQ, Deng ZR, Hong DY. 1998. The systematic position of Beesia: evidence from ITS (nrDNA) sequence analysis. J Syst Evol. 36(5):403–410.
- Xiao PG. 1979. Cimicifuga L. In: Anonymous, editor, Flora Reipublicae Popularis Sinicae. Vol. 27. Beijing: Science Press; p. 93–103.
- Yang QE, Gu Z, Sun H. 1995. The karyotype of Beesia deltophylla and its systematic significance. J Syst Evol. 33(3):225–229.
- Zhai W, Duan XS, Zhang R, Guo C, Li L, Xu G, Shan HY, Kong HZ, Ren Y. 2019. Chloroplast genomic data provide new and robust insights into the phylogeny and evolution of the Ranunculaceae. Mol Phylogenet Evol. 135:12–21.