Abstract
Artocarpus gomezianus is a medicinal species native to Asia. To infer its phylogenetic relationship to the other Moraceae, the complete chloroplast genome of A. gomezianus was sequenced. The whole chloroplast genome is 160,743 bp in length, consisting of a pair of inverted repeat (IR) regions of 25,691 bp, one large single-copy (LSC) region of 89,241 bp, and one small singlecopy (SSC) region of 20,120 bp. The overall GC content of the complete chloroplast genome is 35.81%. Maximum likelihood analysis using 11 complete plastomes of the Moraceae and Cannabis sativa (Cannabaceae) designated as the outgroup, resolved A. gomezianus in a clade with A. petelotii and A. hypargyreus. These phylogenetic results are not consistent with previous findings based on nuclear loci in which A. gomezianus was grouped as a sister to a clade containing A. petelotii and A. hypargyreus. The complete chloroplast genome of A. gomezianus will provide a powerful tool to accelerate pharmacological development, systematics, and future phylogenetic studies in the Moraceae.
Artocarpus gomezianus Wall. ex Trécul is a plant species with a variety of medicinal values, among approximately 70 other species classified to the genus Artocarpus (Moraceae) (Williams et al. Citation2017). The arylbenzofurans, flavonoids, phenolics and stilbenoids extracted from A. gomezianus are α-glucosidase inhibitors (Nuntawong et al. Citation2019) and tyrosinase inhibitors (Likhitwitayawuid et al. Citation2000; Likhitwitayawuid and Sritularak Citation2001), show antiherpetic activities (Likhitwitayawuid et al. Citation2005, Citation2006). A facile and eco-friendly combustion method mediated by A. gomezianus fruit was applied to synthesize spherical nanoparticles of zinc oxide, which were used as anticancer, antibacterial and antifungal materials (Anitha et al. Citation2018). No genomic resources have been reported for this species, and this hinders the development of A. gomezianus pharmacological properties. In the present study, we report the complete chloroplast genome sequence of A. gomezianus to contribute to further phylogenetic and population genetic studies.
The voucher specimen of A. gomezianus was collected from Menglun Town, Mengla County of Xishuangbanna (Yunnan, China; Long. 101.2742 E, Lat. 21.9198 N; Altitude: 555.35 m), and is deposited at the herbarium of South China Botanical Garden, Chinese Academy of Sciences (contact person: Shuangwen Deng, email: [email protected], accession number: SCBG-BN-26). The DNA was extracted from fresh young leaves of A. gomezianus following the modified CTAB-chloroform protocol (Doyle and Doyle Citation1990). The DNA was sequenced on an Illumina Hiseq 2000 platform at Novogene-Beijing (Illumina, San Diego, CA) and generated 4.5 Gb raw data. The chloroplast genome sequence was assembled using the default settings in NOVOPlasty (Dierckxsens et al. Citation2017). A ribulose-1, 5-bisphosphate carboxylase/oxygenase (rbcL) gene sequence from A. heterophyllus (GenBank accession no. MK303549) served as the seed sequence, and the complete chloroplast genome sequence of A. heterophyllus was used as a reference to resolve the inverted repeat in the chloroplast genome of A. gomezianus. The assembled chloroplast genome was annotated using PGA (Qu et al. Citation2019) and GeSeq (Tillich et al. Citation2017), and also adjusted manually. The annotated chloroplast genomic sequence was deposited in GenBank under the accession number: MW837773.
The chloroplast genome of A. gomezianus is 160,743 bp in size, displays a 35.81% GC content, and includes a characteristic quadripartite structure with a LSC of 89,241 bp, an SSC of 20,120 bp and a pair of IRs of 25,691 bp. A total of 131 genes were identified, including 85 protein-coding, 38 tRNA and 8 rRNA genes. The chloroplast genome length of A. gomezianus is 266 bp, 209 bp smaller than that of A. petelotii (161,009 bp, MW250918) (Chen and Liu Citation2021) and A. hypargyreus (160,952 bp, MN720648) (Li et al. Citation2020), but 356 bp larger than that of A. heterophyllus (160,387 bp, MG434693) (Liu et al. Citation2017). The GC content and genome organization of A. gomezianus is similar to that of A. petelotii and A. hypargyreus.
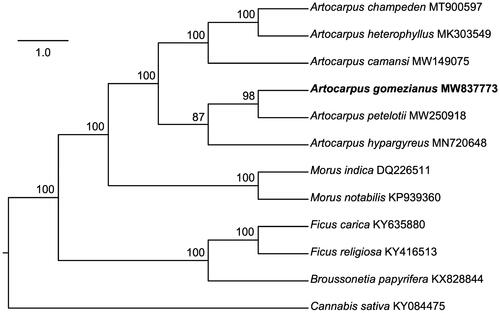
To confirm the phylogenetic position of A. gomezianus, the complete plastome sequences of ten previously published Moraceae species, including A. camansi, A. champeden, A. heterophyllus, A. hypargyreus, A. petelotii, Morus indica, M. notabilis, Ficus carica, F. religiosa and Broussonetia papyrifera, and one outgroup Cannabis sativa (Cannabaceae) were downloaded from the NCBI GenBank database. Complete sequences were aligned using the default settings in MUSCLE (Edgar Citation2004), and the phylogenetic tree () was constructed using the TVM + F+R2 model in IQ-TREE (Nguyen et al. Citation2015). Based on 1000 bootstrap replicates, the phylogenetic tree strongly supported that A. gomezianus is placed in a clade with A. petelotii and A. hypargyreus (). However, in the phylogenetic framework constructed from 517 genes, A. gomezianus was sister to the clade containing A. petelotii and A. hypargyreus (Gardner et al. Citation2021). It suggests that the chloroplast of A. gomezianus may share close ancestry with that of A. petelotii, although they are clearly distinct in nuclear DNA sequences. In conclusion, the A. gomezianus chloroplast genome reported here will provide a solid foundation for phylogenetic and evolutionary studies in Artocarpus and may improve the detection of various molecular mechanisms of the medicinal properties found in A. gomezianus.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The data is available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/, under the accession number [MW837773]. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA717896, SRS8590981, and SAMN18515340 respectively.
Additional information
Funding
References
- Anitha R, Ramesh KV, Ravishankar TN, Sudheer Kumar KH, Ramakrishnappa T. 2018. Cytotoxicity, antibacterial and antifungal activities of ZnO nanoparticles prepared by Artocarpus gomezianus fruit mediated facile green combustion method. J Sci Adv Mater Devices. 3(4):440–451.
- Chen HH, Liu Q. 2021. The plastid genome of a narrowly distributed species Artocarpus petelotii (Moraceae). Mitochondrial DNA B Resour. 6 (2):454–455.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Doyle JJ, Doyle JL. 1990. Isolation of plant DNA from fresh tissue. Focus. 12:13–15.
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797.
- Gardner EM, Johnson MG, Pereira JT, Puad ASA, Arifiani D, Wickett NJ, Zerega NJC. 2021. Paralogs and off-target sequences improve phylogenetic resolution in a densely sampled study of the breadfruit genus (Artocarpus, moraceae). Syst Biol. 70(3):558–575.
- Li DL, Gul J, He JM. 2020. The complete chloroplast genome sequence of Artocarpus hypargyreus. Mitochondrial DNA B Resour. 5 (1):910–911.
- Likhitwitayawuid K, Chaiwiriya S, Sritularak B, Lipipun V. 2006. Antiherpetic flavones from the heartwood of Artocarpus gomezianus. Chem Biodivers. 3(10):1138–1143.
- Likhitwitayawuid K, Supudompol B, Sritularak B, Lipipun V, Rapp K, Schinazi RF. 2005. Phenolics with anti-HSV and anti-HIV activities from Artocarpus gomezianus, Mallotus pallidus, and Triphasia trifolia. Pharm Biol. 43(8):651–657.
- Likhitwitayawuid K, Sritularak B. 2001. A new dimeric stilbene with tyrosinase inhibitiory activity from Artocarpus gomezianus. J Nat Prod. 64(11):1457–1459.
- Likhitwitayawuid K, Sritularak B, De-Eknamkul W. 2000. Tyrosinase inhibitors from Artocarpus gomezianus. Planta Med. 66(3):275–277.
- Liu J, Niu Y-F, Ni S-B, Liu Z-Y, Zheng C, Shi C. 2017. The complete chloroplast genome of Artocarpus heterophyllus (Moraceae). Mitochondrial DNA B Resour. 3(1):13–14.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Nuntawong P, Kongkatitham V, Likhitwitayawuid K, Mekboonsonglarp W, Sukrong S, Tanasupawat S, Sritularak B. 2019. New 2-arylbenzofurans from the root bark of Artocarpus gomezianus and their α-glucosidase inhibitory activity. Nat Prod Res. 33(10):1436–1441.
- Qu XJ, Moore MJ, Li DZ, Yi TS. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15:50.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq - versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Williams EW, Gardner EM, Harris R, III, Chaveerach A, Pereira JT, Zerega NJC. 2017. Out of Borneo: biogeography, phylogeny and divergence date estimates of Artocarpus (Moraceae). Ann Bot. 119(4):611–627.