Abstract
Cycas bifida (Dyer) K.D.Hill (2004) is an extremely small population-protected species of China. In this study, we reported the first chloroplast genome sequence of C. bifida. The chloroplast genome of C. bifida included two single-copy regions (large single-copy (LSC) and small single-copy (SSC)) and a pair of inverted repeats (IRs) regions comprising 88,946 bp, 23,107 bp, and 25,053 bp, respectively. The complete chloroplast genome of C. bifida contains 131 genes, including 86 protein-coding genes, 37 transfer RNA genes, and 8 ribosomal RNA genes. The overall GC content of the C. bifida chloroplast genome is 39.41%, and the LSC, SSC, and IR regions occupy 38.70%, 36.52%, and 42.02%, respectively. A phylogenetic analysis was performed based on complete chloroplast genomes from 15 species and found that C. bifida was closely related to Cycas szechuanensis W.C.Cheng & L.K.Fu.
Cycas bifida (Dyer) K.D.Hill (2004), an extremely small population-protected species of China, belongs to the cycad family (Cycadaceae, Cycas) (Zheng et al. 2018; Lin et al. Citation2019). Cycads are regarded as the ‘Living Fossils’ and belong to a specialized group of plants having ancient lineage possessing great significance from the evolutionary point of view (Goel and Khuraijam Citation2015). Cycas bifida was listed as rare and endangered plant (Ying Citation2013), with a geographical range restricted to a total of two known countries (Vietnam and China) (Osborne Citation1995). However, the complete chloroplast genome of C. bifida has not been sequenced.
In this study, we assembled and characterized the complete chloroplast (cp) genome of C. bifida and determined its molecular phylogeny and genetic information. Fresh leaf tissues of C. bifida were collected from Guangxi Institute of Botany, Guilin, China (GPS: 25°4’44” N, 110°18’1” E). Its voucher specimen was deposited in the herbarium at the Guangxi Institute of Botany (http://www.gxib.cn/spIBK/, Z. C. Lu, email: [email protected]) with the voucher number as IBK00435048. Total genomic DNA was extracted by the modified CTAB method (Doyle Citation1991). A total of 6 G raw data from Illumina Hiseq Platform were screened and assembled into a complete chloroplast genome by GetOrganelle in a typical way (Jin et al. Citation2020). The assembled sequences were annotated using the tool CPGAVAS2 and using Cycas szechuanensis (NC_042668.1) as reference (Shi et al. Citation2019, Wang et al. Citation2018). The annotated chloroplast genome sequence was submitted to the NCBI GenBank database under the accession number MW900434.
Based on the assembling results, the length of the complete chloroplast genome of C. bifida is 162,159 bp. The chloroplast genome of C. bifida included two single-copy regions (large single-copy (LSC) and small single-copy (SSC)) and a pair of inverted repeats (IRs) regions comprising 88,946 bp, 23,107 bp, and 25,053 bp, respectively. The complete chloroplast genome of C. bifida contains 131 genes, including 86 protein-coding genes, 37 transfer RNA genes, and 8 ribosomal RNA genes. The overall GC content of C. bifida chloroplast genome is 39.41%, and the LSC, SSC, and IR regions occupy 38.70%, 36.52%, and 42.02%, respectively.
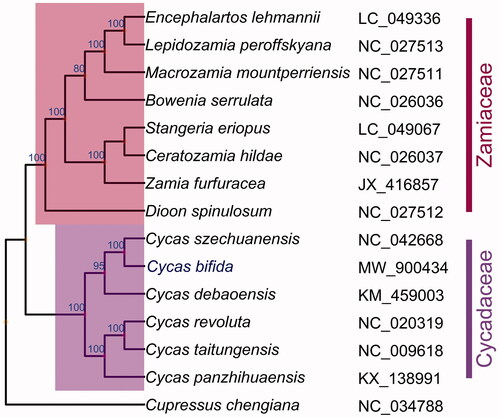
To reveal the phylogenetic position of C. bifida with other Cycads, the complete chloroplast genomes of 14 published species within Cycadales and one outgroup Cupressus chengiana (NC_034788.1) were downloaded from the NCBI. A total of 83 coding sequences (accD, atpA, atpB, atpE, atpF, atpH, atpI, ccsA, cemA, chlB, chlL, chlN, clpP, infA, matK, ndhA, ndhB, ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, ndhK, petA, petB, petD, petG, petL, petN, psaA, psaB, psaC, psaI, psaJ, psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbT, psbZ, rbcL, rpl14, rpl16, rpl2, rpl20, rpl22, rpl23, rpl32, rpl33, rpl36, rpoA, rpoB, rpoC1, rpoC2, rps11, rps12, rps14, rps15, rps16, rps18, rps19, rps2, rps3, rps4, rps7, rps8, ycf1, ycf12, ycf2, ycf3, and ycf4) were extracted, and aligned by using MUSCLE (Edgar Citation2004). The maximum likelihood (ML) tree with 100 bootstrap replicates was performed with RaxML v 8.2.12 (Stamatakis Citation2014). From the ML phylogenetic tree, C. bifida was closely related to Cycas szechuanensis. The complete chloroplast genome of C. bifida will provide useful genetic resources for further studying on genetic diversity of this important species and help us protect this extremely endangered plant ().
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MW900434. The associated BioProject, SRA, and BioSample numbers are PRJNA723747, SRX10656572, and SAMN18828092 respectively. Treefile of 15 species and genes for phylogenetic analysis were deposited at Figshare: https://doi.org/10.6084/m9.figshare.14784537.v5.
Additional information
Funding
References
- Doyle J. 1991. DNA protocols for plants. Vol. 57. In: Hewitt GM, Johnston AWB, Young JPW, editors. Molecular techniques in taxonomy. Switzerland: Springer; p. 283–293.
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32(5):1792–1797.
- Goel AK, Khuraijam J. 2015. Cycads: an overview. In: Bahadur B, Rajam MV, Sahijram L, Krishnamurthy KV, editors. Plant biology and biotechnology. New Delhi, India: Springer; p. 349–360.
- Jin J-J, Yu W-B, Yang J-B, Song Y, Depamphilis CW, Yi T-S, Li D-Z. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241–231.
- Lin J, Li J, Liang R. 2019. Intraspecific and interspecific competition of Cycas micholitzii in the shrub layer. J Forest Environ. 39:159–164.
- Osborne R. 1995. The world cycad census and a proposed revision of the threatened species status for cycad taxa. Biol Conserv. 71(1):1–12.
- Shi L, Chen H, Jiang M, Wang L, Wu X, Huang L, Liu C. 2019. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47(W1):W65–W73.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Wang Y, Wang Z, Zhang L, Yang X. 2018. The complete chloroplast genome of Cycas Szechuanensis, an extremely endangered species. Mitochondrial DNA Part B. 3(2):974–975.
- Ying H. 2013. Rare and endangered plants in China. Beijing: China Forestry Publishing House.
- Zheng Y, Chiang T-Y, Huang C-L, Gong X. 2018. Highly diverse endophytes in roots of Cycas bifida (Cycadaceae), an ancient but endangered gymnosperm. J Microbiol. 56(5):337–345.