Abstract
Dichroa febrifuga, seen as a medicinal plant, has a long history in traditional Chinese medicine. In this study, we adopted Illumina Hiseq sequencing technology in order to determine the first complete chloroplast (cp) genome of D. febrifuga. The cp genome was 157,647 bp in length, including a large single-copy (LSC) region of 86,728 bp, a small single-copy (SSC) region of 18,675 bp, and a pair of inverted repeat (IR) regions of 26,122 bp. The genome encoded 128 genes, including 84 protein-coding genes, 36 tRNA genes, and 8 rRNA genes. The phylogenetic analysis based on 20 complete cp genome sequences revealed that D. febrifuga was the sister of the ancestor of the reported Hydrangeeae species. The findings of the study will serve as a stepping stone for follow-up researches regarding the development of the D. febrifuga species.
Dichroa febrifuga Lour., a member of the genus Dichroa in Hydrangeaceae, grows in grove-shaded and humid mountainous areas, mainly distributing in Sichuan, Guizhou, Hunan, and Hubei of China. According to the ‘Pharmacopoeia of China’ (2020 version), D. febrifuga is one of the common Chinese herbal medicines in the folk, which can be used to treat malaria, and eliminate phlegm. At the same time, D. febrifuga is rich in quinazolone alkaloids, coumarins, steroids, polyphenols and other chemical components (Zhang et al. Citation2010). It not only has the effects of anti-malaria, anti-tumor, anti-insect (Han Citation2021), promoting wound healing and other pharmacological effects (Li et al. Citation2011), but is also a traditional Chinese medicine raw material used for developing antiparasitic drugs in veterinary medicine (Chen et al. Citation2021). However, as an important medicinal plant, there is no genomic information that has been reported so far. In this study, we reported and characterized the first complete chloroplast (cp) genome of D. febrifuga based on Illumina Hiseq sequencing.
Fresh leaves of D. febrifuga were collected from Nanchuan, chongqing, China (107°21′E, 29°13′N, 591 m). The voucher specimen was conserved in Chongqing Institute of Medicinal Plant Cultivation (accession number: CIMPC-RFM-20210303, Contact person: Fengming Ren; Email: [email protected]). In the experiment, the high-quality whole genomic DNA was extracted by the kit method (Beijing Kinco Biological Company). The purity and integrity of the extracted DNA were analyzed by Nanodrop (Thermo Fisher Scientific) and agarose gel electrophoresis. High quality DNA was used to generate paired-end libraries with insert size of 350 bp and about 2.94 Gb raw reads were generated by Illumina Hiseq 2500 Platform (Illumina, Hayward, CA, USA). The raw data from the platform was removed low-quality reads and adapters by trimmomatic (version 0.35) with default paprameters (Bolger et al. Citation2014). Using the clean data with 150 bp paired-end read length obtained from the raw data, a cp genome was assembled by NOVOPlasty (version 4.1) with the default parameters (Nicolas et al. Citation2016) and was annotated by CPGAVAS2 with default parameters (http://47.96.249.172:16019/analyzer/home) (Shi et al. Citation2019). After manual check and adjustment, the annotated cp genome was submitted to the GenBank (MW928534).
The complete D. febrifuga cp genomes was 157,647 bp in size and exhibited a typical angiosperm cp circular structure, containing four regions: large single-copy region (LSC: 86,728 bp), small single-copy region (SSC: 18,675 bp), and a pair of inverted repeat regions (IR: 26,122 bp). The GC contents of whole genome and each region of the genome were 37.86% (whole genome), 36.05% (LSC region), 31.68% (SSC region), and 43.09% (IR region). The GC content accorded with the typical characteristics of angiosperm cp genome. The cp genome encoded 128 functional genes, including 84 protein-coding genes, 36 tRNA genes, and 8 rRNA genes.
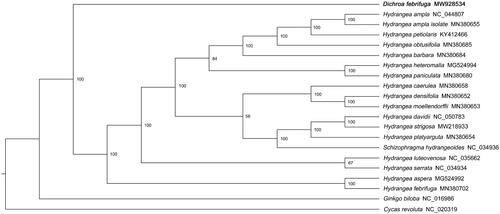
To reveal the phylogenetic relationship of D. febrifuga within Hydrangeeae, additional 19 species of Hydrangeeae from the NCBI website were selected to study. With Ginkgo biloba (NC_016986.1) and Cycas revoluta (NC_020319.1) as outgroups, a phylogenetic tree was built by using the 21 cp genome sequences. The 21 sequences were compared by MAFFT software (version 7.487) with the parameter of ‘–auto’ (Katoh and Standley Citation2013). Based on the aligned sequences, a Maximum-likelihood phylogenetic tree was built with 1000 bootstrap replicates by IQ-TREE (version 1.6.12) (Lam-Tung et al. Citation2015) under parameters of ‘-nt AUTO -m MFP -bb 1000 –bnni’. Phylogenetic analysis showed that D. febrifuga was the sister of the ancestor of the reported Hydrangeeae species. ().
Acknowledgments
We thank Professor Qiuping Tan for Providing plant material.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, under the accession number MW928534. The associated Bio-Project, Bio-Sample and SRA numbers are PRJNA543381, SAMN19416297 and SRR14688233, respectively.
Additional information
Funding
References
- Bolger AM, Marc L, Bjoern U. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120.
- Chen DZ, Sun JH, Zhang JY, Zhou XZ. 2021. Discussion on the research and development direction of antiparasitic drugs for animals. J Trad Chinese Vet Med. 40(02):35–40.
- Han JH. 2021. Study on the prevention and treatment of broiler coccidiosis with Chinese herbal medicine Dichroa febrifuga. China Animal Health. 23(04):4–42.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Lam-Tung N, Schmidt HA, Arndt VH, Bui QM. 2015. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Li Y, Liu MC, Jin LH, Hu DY, Sy Y. 2011. Research progress of Dichroa febrifuga chemical composition and biological activity. Guangzhou Chemical Industry. 39(09):7–9.
- Nicolas D, Patrick M, Guillaume S. 2016. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Shi L, Chen H, Jiang M, Wang L, Wu X, Huang L, Liu C. 2019. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47(W1):W65–73.
- Zhang Y, Li C, Lei GL. 2010. Study on the chemical constituents of Dichroa febrifuga. Chin J Exp Form. 16(05):40–42.