Abstract
The file ramshorn snail Planorbella pilsbryi Baker, 1926 (Gastropoda: Hygrophila: Planorbidae) is a widespread herbivorous North American freshwater snail found in diverse habitats, including standing and moving water bodies. Genome skimming by Illumina sequencing allowed the assembly of a complete nuclear rRNA repeat sequence and a complete circular mitogenome of 13,720 bp from P. pilsbryi consisting of 75.3% AT nucleotides, 22 tRNAs, 13 protein-coding genes, 2 rRNAs and a control region in the typical order found in panpulmonate snails. Planorbella pilsbryi COXI features a rare TTG start codon while COXII, CYTB, ND2, ND3, and ND5 exhibit incomplete stop codons completed by the addition of 3′ A residues to the mRNA. Phylogenetic reconstruction of mitochondrial protein-coding gene and rRNA sequences places P. pilsbryi as sister taxon to Planorbella duryi (Planorbidae) within family Planorbidae, which is consistent with previous phylogenetic hypotheses.
The file ramshorn snail, Planorbella pilsbryi Baker 1926 is a common freshwater snail species native to southern Canada and the United States (Baker Citation1945; Clarke Citation1981; McKillop Citation1996). These snails play vital roles in a variety of environments as principal grazers, contributors to nutrient cycling, and prey for invertebrate and vertebrate predators (Baker Citation1945; Johnson et al. Citation2013). Furthermore, they are used in toxicology studies to determine the effects of herbicides and insecticides on freshwater invertebrate communities (Prosser et al. Citation2016). However, taxonomic questions have been raised that may have implications for interpretation of ecological studies. For example, some taxonomists suggest that P. pilsbryi is a genetic variant and eco-phenotype of Planorbella trivolvis, rather than a distinct species (Pip Citation1987). To better understand the taxonomy and evolutionary ecology of P. pilsbryi, we characterized its mitogenome and nuclear rRNA repeat.
Here we report the complete mitochondrial genome sequence of P. pilsbryi from specimen PlPi_MW889961, collected from the Marais River in Manitoba, Canada (GPS 49.127 N, 97.303 W) on 11 June 2019 that was deposited in the collection of the Manitoba Museum (https://manitobamuseum.ca/collections-research/, Randall Mooi, [email protected]), voucher MM 67397. The specimen was identified as P. pilsbryi using criteria outlined by Baker (Citation1945), namely a wide, bowl-like spire, a deeply excavated umbilical region, and a shell height equal to roughly 50% of the shell’s diameter. DNA was extracted from 30 mg of tissue from the head-foot region using an E.Z.N.A Mollusk DNA kit (Omega Bio-tek Inc., Norcross, USA) before it was sheared by sonication and a fragment library was prepared using the NEBNext Ultra II DNA Library Prep Kit for Illumina (New England Biolabs, Ipswich, Massachusetts) as previously described (Peters and Marcus Citation2017). Finally, it was sequenced by Illumina NovaSeq6000 (San Diego, California, USA) at Genome Quebec.
Mitogenome assembly of P. pilsbryi (Genbank accession MW889961) was performed by mapping the resulting sequence library of 21,753,838 paired 150 bp reads (Genbank SRA PRJNA718624) to a Planorbella duryi reference mitogenome (Gastropoda: Planorbidae, KY514384 (Schultz et al. Citation2018)) using 5 iterations of the medium sensitivity settings of Geneious 2020.2 as described by Marcus (Citation2018). Annotation was in reference to P. duryi and Biomphalaria choanomphala (Hygrophila: Planorbidae, MG431964) mitogenomes (Zhang et al. Citation2018). The complete P. pilsbryi nuclear rRNA repeat (8182 bp) (MW883067) was also assembled and annotated using reference sequences from the 18S rRNA gene from Galba cubensis (Gastropoda: Lymnaeidae, Z83831) (Bargues et al. Citation1997); the partial 18S rRNA, ITS1, 5.8S rRNA, ITS2, and partial 28S rRNA region from Satsuma polymorpha (Gastropoda: Camaenidae, AB597368) (Hoso et al. Citation2010); the 28S rRNA genes from P. trivolvis (KY319366) (unpublished) and Pomacea bridgesi (Gastropoda: Ampullariidae, DQ279984) (Giribet et al. Citation2006); and the complete rRNA repeat from Macrosoma conifera (Lepidoptera: Hedylidae, MT878224) (McCullagh et al. Citation2020).
The P. pilsbryi circular 13,720 bp mitogenome assembly was composed of 5,620 paired reads with nucleotide composition: 33.8% A, 11.4% C, 13.3% G, and 41.5% T. The gene composition and order in P. pilsbryi is identical to all known panpulmonate snails except for Physella acuta (Physidae) (Nolan et al. Citation2014). Protein-coding gene start codons in the P. pilsbryi mitogenome include ATG (COXIII, ND1, ND3, and ND5), ATT (ATP6, ATP8, and ND2), ATA (COXII, CYTB, ND4, and ND6), ATC (ND4L), and TTG (COXI). The mitogenome contains 7 protein-coding genes (ATP6, ATP8, COXI, COXIII, ND1, ND4, and ND6) with TAA stop codons, 1 protein-coding gene (ND4L) with a TAG stop codon, and 5 protein-coding genes (COXII, CYTB, ND2, ND3, and ND5) with single-nucleotide (T) stop codons completed by post-transcriptional addition of 3′ A residues. The locations and structures of tRNAs were determined using ARWEN v.1.2 (Laslett and Canback Citation2008). All 22 tRNAs have typical cloverleaf secondary structures except for: trnG (TCC) where the dihydrouridine arm (D-arm) is replaced by a loop (D-replacement loop), as well as trnS1 (TGA), trnS2 (GCT), and trnLA (TTT), where the TφC (T) arm and variable (V) loop are replaced by a loop (TV-replacement loop). The nuclear rRNA sequences increase the number of genetic loci that are available for future use in phylogenetic analyses of the Planorbidae.
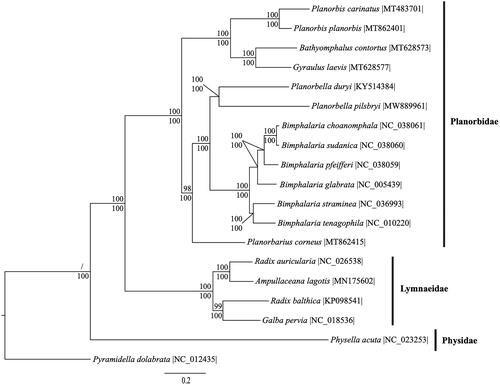
We reconstructed a phylogeny using 11 protein-coding genes and 2 rRNA genes from P. pilsbryi, 17 gastropod species within the superorder Hygrophila, and the gastropod Pyramidella dolabrata (superfamily Pyramidelloidea) as an outgroup (Grande et al. Citation2008). Two protein-coding genes (ATP6 and ATP8) were excluded to increase taxon sampling in the phylogeny. Mitogenome sequences were aligned in CLUSTAL Omega (Sievers et al. Citation2011). The best fit model of evolution was GTR + I + G according to jModeltest 2.1.7 (Darriba et al. Citation2012). For maximum likelihood analysis, we employed the GTRGAMMA model and calculated bootstrap probabilities with 1,000 replicates in RaxML v8 (Stamatakis Citation2014). For Bayesian inferences, 2 independent analyses were run with 4 chains each for 10,000,000 generations with samples taken every 1,000 generations using MrBayes v3.2.6 (Ronquist and Huelsenbeck Citation2003). A burn-in of 2,500 was used and the remaining trees were used to calculate the Bayesian posterior probabilities. Phylogenetic analysis supports the monophyletic intrageneric relationship between P. duryi and P. pilsbryi (). The mitogenome of P. pilsbryi can be used to clarify the evolutionary relationships among gastropod taxa at higher (family) and lower (species) levels of taxonomy. Further, this resource can help ground ecological investigations that include Planorbella species in a clearer taxonomic framework.
Figure 1. Phylogenetic reconstruction of mitogenomes (11 protein-coding genes and 2 rRNA genes) of Planorbella pilsbryi, 17 additional sequences from superorder Hygrophila, and 1 outgroup species, Pyramidella dolbrata, from superfamily Pyramidelloidea. Sequences were analyzed using maximum likelihood (ML) and Bayesian inference (BI). The Bayesian inference tree is shown. Numbers above each node are ML bootstrap values, while Bayesian posterior probabilities are displayed under each node.
Acknowledgements
We thank Carly Gair for assistance with DNA extraction. We thank Genome Quebec for assistance with library preparation and sequencing.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at [https://www.ncbi.nlm.nih.gov] (http://www.ncbi.nlm.nih.gov/) under the accession nos. MW889961 and MW883067. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA718624, SRX10533846, and SAMN18543613. The specimen was deposited in the Manitoba Museum (https://manitobamuseum.ca/collections-research/, Randall Mooi, [email protected]), voucher MM 67397.
Additional information
Funding
References
- Baker FC. 1945. The molluscan family Planorbidae. Urbana, IL: University of Illinois Press.
- Bargues MD, Mangold AJ, Munoz-Antoli C, Pointier JP, Mas-Coma S. 1997. SSU rDNA characterization of lymnaeid snails transmitting human fascioliasis in South and Central America. J Parasitol. 83(6):1086–1092.
- Clarke AH. 1981. The freshwater molluscs of Canada. Ottawa, Canada: National Museum of Natural Sciences, National Museums of Canada.
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. JModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9(8):772.
- Giribet G, Okusu A, Lindgren AR, Huff SW, Schrodl M, Nishiguchi MK. 2006. Evidence for a clade composed of molluscs with serially repeated structures: monoplacophorans are related to chitons. Proc Natl Acad Sci USA. 103(20):7723–7728.
- Grande C, Templado J, Zardoya R. 2008. Evolution of gastropod mitochondrial genome arrangements. BMC Evol Biol. 8:61.
- Hoso M, Kameda Y, Wu SP, Asami T, Kato M, Hori M. 2010. A speciation gene for left-right reversal in snails results in anti-predator adaptation. Nat Commun. 1(133):133.
- Johnson PD, Bogan AE, Brown KM, Burkhead NM, Cordeiro JR, Garner JT, Hartfield PD, Lepitzki DAW, Mackie GL, Pip E, et al. 2013. Conservation status of freshwater gastropods of Canada and the United States. Fisheries. 38(6):247–282.
- Laslett D, Canback B. 2008. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 24(2):172–175.
- Marcus JM. 2018. Our love-hate relationship with DNA barcodes, the Y2K problem, and the search for next generation barcodes. AIMS Genet. 5(1):1–23.
- McCullagh BS, Alexiuk MR, Payment JE, Hamilton RV, Lalonde MML, Marcus JM. 2020. It's a moth! It's a butterfly! It's the complete mitochondrial genome of the American moth-butterfly Macrosoma conifera (Warren, 1897) (Insecta: Lepidoptera: Hedylidae)! Mitochondr DNA B Resour. 5(3):3633–3617.
- McKillop B. 1996. Geographic and environmental distribution of freshwater gastropods in Manitoba, Canada. Vol. 1. Manitoba Museum of Man and Nature Occasional Series. Winnipeg: Manitoba Museum of Man and Nature. p. 1–34.
- Nolan JR, Bergthorsson U, Adema CM. 2014. Physella acuta: atypical mitochondrial gene order among panpulmonates (Gastropoda). J Molluscan Stud. 80(4):388–399.
- Peters MJ, Marcus JM. 2017. Taxonomy as a hypothesis: testing the status of the Bermuda buckeye butterfly Junonia coenia bergi (Lepidoptera: Nymphalidae). Syst Entomol. 42(1):288–300.
- Pip E. 1987. Ecological differentiation within genus Helisoma in Central Canada. Nautilus. 101(1):33–44.
- Prosser RS, de Solla SR, Holman EAM, Osborne R, Robinson SA, Bartlett AJ, Maisonneuve FJ, Gillis PL. 2016. Sensitivity of the early-life stages of freshwater mollusks to neonicotinoid and butenolide insecticides. Environ Pollut. 218:428–435.
- Ronquist F, Huelsenbeck JP. 2003. MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19(12):1572–1574.
- Schultz JH, Bansbach LM, Bremmer JA, Dimmler KM, Forde QA, Gagliano EM, Glenn EM, Greengrass CM, Hayes JP, Kraus AL, et al. 2018. The mitochondrial genome of the planorbid snail Planorbella duryi. Mitochondrial DNA B Resour. 3(2):972–973.
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Weizhong L, Lopez R, McWilliam H, Remmert M, Söding J, et al. 2011. Fast, scalable generation of high quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 7(539):539.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Zhang S-M, Bu L, Laidemitt MR, Lu L, Mutuku MW, Mkoji GM, Loker ES. 2018. Complete mitochondrial and rDNA complex sequences of important vector species of Biomphalaria, obligatory hosts of the human-infecting blood fluke, Schistosoma mansoni. Sci Rep. 8(1):7341.
