Abstract
The complete mitogenome of Cochylidia moguntiana (Rössler, 1864) was sequenced and analyzed. The genome is 15,433 bp long with a high A + T content (80.6%), and consists of 13 protein-coding genes, 22 tRNA genes, 2 rRNA genes, and a noncoding control region. A phylogenetic analysis of 18 tortricid species for which mitogenes are available showed strong support for the monophyly of Tortricinae.
Since the complete mitochondrial genome of the smaller tea tortrix, Adoxophyes honmai was reported in 2006, 20 complete mitochondrial genomes of Tortricidae have been published (Lee et al. Citation2006; Qi et al. Citation2021). The first complete mitochondrial genome of the tribe Cochylini (Tortricidae), that of Cochylimorpha cultana (Lederer, 1855), was only recently reported by Qi et al. (Citation2021). We sequenced the mitochondrial genome of C. moguntiana (Lederer, 1855), a second species of the tribe Cochylini, which provided additional molecular data for an analyses of phylogenetic and evolutionary relationships within Tortricidae.
Cochylidia moguntiana is a member of one of the largest genera in the tribe Cochylini (Tortricidae: Tortricinae). A recent molecular phylogenetic analysis of the tribe (Brown et al. Citation2019) placed Cochylimorpha as sister to Eugnosta Hübner, [1825] 1816). At present, Cochylimorpha comprises 97 species worldwide (Gilligan et al. Citation2018), with greatest species richness in the Palearctic – China, Russia, and Europe in particular (Sun and Li Citation2013). The larvae of Cochylimorpha utilize mainly Artemisia species (Asteraceae), feeding on the seeds, stems, and roots (Razowski Citation1987). Numerous species of Cochylimorpha (or Artemisia?) are found in open, dry biotopes, including the steppes (Razowski Citation2009).
An adult male of C. moguntiana was collected in Funing County, Hebei Province, China (39.50°N, 119.26°E) in 2018 by light trap. The specimen was identified based on the text and images in Razowski (Citation1970) and Sun and Li (Citation2012). Total genomic DNA was extracted from the muscle tissues of the legs of the adult using the DNeasy Blood and Tissue kit (QIAGEN Sciences, Valencia, CA, USA). The DNA and voucher specimen of C. moguntiana are deposited in the Insect Collection, College of Life Sciences, Dezhou University, Shandong, China (Yinghui Sun, [email protected]), under the accession code DZU002. COI of C. moguntiana (GenBank accession: MM17263) was used as bait to extract the mitogenome of C. moguntiana. The genomic DNA was subsequently pooled with that of other insect species and sequenced using the Illumina Nova6000 (PE150, Illumina, San Diego, CA, USA) platform at Novogene Co., Ltd. (Beijing, China). The software IDBA-1.1.1 (Peng et al. Citation2012) was employed to assemble the data, with similarity set at 0.98. The mitogenome of C. moguntiana was then extracted using a Blast search (Altschul et al. Citation1990) with COI as the bait sequence (Crampton-Platt et al. Citation2015), and the percentage of identical matches was 100%. The mitogenome annotation was conducted using the methods of Zheng et al. (Citation2020).
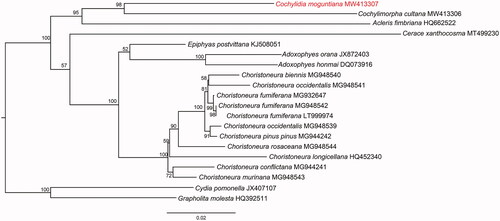
Mitochondrial genomes of 18 additional Tortricinae species available from GenBank were used for a phylogenetic analysis. The sequences were concatenated with alignments of 13 PCGs using the default settings in MAFFT (Katoh and Standley Citation2013). Maximum-likelihood (ML) reconstruction was performed using IQ-TREE (Nguyen et al. Citation2015) with 1000 bootstraps replicates and the PMSF acid substitution model. Two species of Olethreutinae, Cydia pomonella (L.) (GenBank accession No. JX407107) and Grapholita molesta (Busck) (GenBank accession No. HQ392511), were used as outgroups.
The mitogenome of C. moguntiana is 15,433 bp in length (GenBank accession No. MW413307), containing 13 protein-coding genes (PCGs), 2 ribosomal RNA genes, 22 transfer RNA genes, and one noncoding control region. The overall nucleotide composition was 40.8% A, 39.8% T, 11.4% C, and 8.0% G, with and A + T content of 80.6%. Most of the 13 PCGs have ATN as the start codon (ATA for ND6; ATC for ATP8; ATG for ATP6, COX2, COX3, CytB, ND1, ND4 and ND4L; ATT for ND2, ND3 and ND5; CGA for COX1). The stop codon TAA is assgned to ATP6, ATP8, COX3, CytB, ND1, ND2, ND3, ND4L, ND6, whereas a single T is assigned to COX1, COX2, ND4 and ND5.
The phylogeny based on the 18 mitogenes from GenBank and our mitogene sequence of C. moguntiana, supports the monophyly of Tortricinae (). Although the combined sequence data do not represent a broad cross-section of the subfamily, the results of the analysis are highly consistent with that of other phylogenetic analyses of the family Tortricidae (e.g. Regier et al. Citation2012).
Acknowledgement
We express our cordial thanks to Johw W. Brown (National Museum of Natural History, Smithsonian Institution, Washington, DC, U.S.A.) for his valuable comments and suggestions to improve the manuscript.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession No. MW413307. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA731101, SRR14598117, SAMN19249501, respectively.
Additional information
Funding
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol. 215(3):403–410.
- Brown JW, Aarvik L, Heikkil M, Brown R, Mutanen M. 2019. A molecular phylogeny of Cochylina, with confirmation of its relationship to Euliina (Lepidoptera: Tortricidae). Syst Entomol. 45(6):1–15.
- Crampton-Platt A, Timmermans MJ, Gimmel ML, Kutty SN, Cockerill TD, Vun Khen C, Vogler AP. 2015. Soup to tree: the phylogeny of beetles inferred by mitochondrial metagenomics of a Bornean rainforest sample. Mol Biol Evol. 32(9):2302–2316.
- Gilligan TM, Baixeras J, Brown JW. 2018. T@RTS: online world catalogue of the Tortricidae (Ver. 4.0). http://www.tortricidae.com/catalogue.asp.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Lee ES, Shin KS, Kim MS, Park H, Cho S, Kim CB. 2006. The mitochondrial genome of the smaller tea tortrix Adoxophyes honmai (Lepidoptera: Tortricidae). Gene. 373:52–57.
- Nguyen LT, Schmidt HA, Haeseler AV, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Peng Y, Leung H, Yiu S, Chin F. 2012. IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics. 28(11):1420–1428.
- Qi ZJ, Sun YH, Sun YL, Wang LY, Xu T. 2021. The complete mitochondrial genome of Cochylimorpha cultana (Lederer, 1855) (Lepidoptera: Tortricidae). Mitochondrial DNA B Resour. 6(9):2507–2509.
- Razowski J. 1987. The genera of Tortricidae (Lepidoptera). Part I: Palaearctic Chlidanotinae and Tortricinae. Acta Zool Cracov. 30(11):141–355.
- Razowski J. 1970. Cochylidae. In: Amsel HG, Gregor F, Reisser, H, editors. Microlepidoptera Palaearctica. Vol. 3. Wien: Verlag Georg Fromme; IV + p. 528, 161.
- Razowski J. 2009. Tortricidae of the Palaearctic region, Volume 2: Cochylini. Kraków-Bratislava: František Slamka; p. 1–195.
- Regier J, Brown J, Mitter C, Baixeras J, Cho S, Cummings M, Zwick A. 2012. A molecular phylogeny for the leaf-roller moths (Lepidoptera: Tortricidae) and its implications for classification and life history evolution. PLOS One. 7(4):e3557417.
- Sun YH, Li HH. 2012. Review of the genus Cochylidia Obraztsov (Lepidoptera: Tortricidae: Cochylini) in China. Zootaxa. 3268(1):1–15.
- Sun YH, Li HH. 2013. A brief summary of tribe Cochylini from China (Lepidoptera: Tortricidae: Tortricinae). Hong Kong Entomol Bull. 5(1):13–18.
- Zheng CG, Ye Z, Zhu XX, Zhang HG, Dong X, Chen P, Bu WJ. 2020. Integrative taxonomy uncovers hidden species diversity in the rheophilic genus Potamometra (Hemiptera: Gerridae). Zool Scr. 49(2):174–186.