Abstract
Camellia semiserrata is a woody plant that produces excellent edible oil and is a common landscaping species in South China. The complete chloroplast genome of C. semiserrata was sequenced, assembled, annotated, and characterized using the Illumina MiSeq platform in this study. The chloroplast genome is 156,968 bp (37.32% GC) and contains a large single copy (LSC) region (86,634 bp), a small single copy (SSC) region (18,272 bp), and a pair of inverted repeat (IR) regions (26,031 bp). It encodes a total of 117 genes, including 81 protein-coding genes, four ribosomal RNA genes, and 32 transfer RNA genes. The phylogenetic tree fully resolved C. semiserrata in a clade with C. reticulata, C. mairei, and C. pitardii. This study contributes to bioinformatics and further phylogeny and conservation studies as well as provides a theoretical basis for the molecular identification of C. semiserrata.
Camellia semiserrata Chi. is mainly distributed in the south of China (Wu et al. Citation2008), belonging to sect. Camellia in the genus Camellia L. (Theaceae) (Ding et al. Citation2012). Due to a long flowering period and the good quality of seed oil, C. semiserrata has both ornamental and economic values and shows great market potential (Rong et al. Citation2021). At present, the phylogenetic history among sect. Camellia species has not yet been fully resolved (Chang and Ren Citation1998; Ming Citation1998). As the chloroplast genome is more conservative than the nuclear genome and mitochondrial genome in terms of genome size, genome structure, and gene content (Palmer Citation1985), the genes obtained from the chloroplast genome have been used to distinguish species and for phylogenomic analysis (Gu et al. Citation2019). Here, we analyzed the chloroplast genome of C. semiserrata to determine its evolutionary systematics and to document the genetic history of the germplasm resources of C. semiserrata.
Young and fresh leaves of C. semiserrata were collected in the National Gene Bank of the Camellia Germplasm Resource (Jiangxi, China; coordinates: 28°44′21.26″N, 115°49′5.42″E), and the specimens were deposited in the Key Laboratory of Camellia Germplasm Conservation and Utilization, Jiangxi Academy of Forestry (contact person: Dong le, [email protected]; specimen voucher: JX20210101). The chloroplast DNA Isolation Kit (Sigma, St. Louis, MO) was used to extract the chloroplast genomic DNA of C. semiserrata following the manufacturer’s instructions. The DNA was sequenced with the Illumina MiSeq using 250 bp paired-end sequencing, then raw data were trimmed with Trimmomatic 0.39 (Bolger et al. Citation2014). Finally, about 1.31 Gb clean data (SRR15294181) were assembled into the complete chloroplast genome with GetOrganelle v1.7.4 pipeline (Jin et al. Citation2020) using the reference genome of C. chekiangoleosa (GenBank accession number MG431968). The genome annotation was performed with GeSeq (Tillich et al. Citation2017), and manually corrected through aligning to the genome annotations of C. chekiangoleosa using Geneious v9.0.2 (http://www.geneious.com). The chloroplast genome of C. semiserrata is deposited in the NCBI database under the accession number MZ403753.
The complete chloroplast genome of C. semiserrata is a circular molecule of 156,968 bp in length, with a large single copy (LSC) region of 86,634 bp, a small single copy (SSC) region of 18,272 bp, and a pair of inverted repeat (IR) regions of 26,031 bp. The total GC content of the chloroplast genome is 37.32%, and the corresponding values of LSC, SSC, and IRs are 35.33%, 30.56%, and 42.99%, respectively. The chloroplast genome encodes 117 unique genes, including 81 protein-coding, 32 tRNA, and four rRNA genes. There are nine protein-coding, four rRNA, and seven tRNA genes repeated in the IR region. In addition, comparative analysis revealed that the chloroplast genome structure and gene locations of C. semiserrata are similar to other related chloroplast genomes of Camellia, such as C. azalea (GenBank accession number KY856741) and C. ptilophylla (GenBank accession number MG797642).
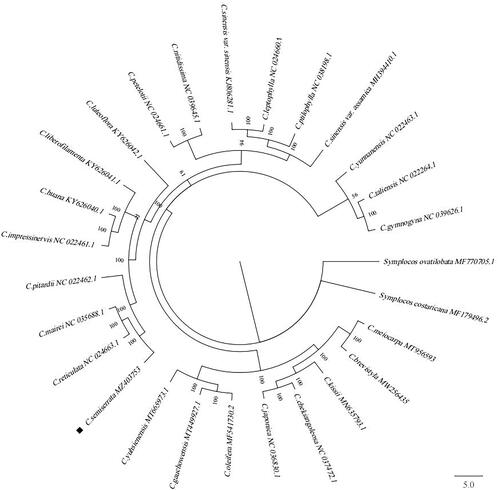
To study evolutionary relationships, the phylogenetic tree was constructed based on chloroplast genome sequences of C. semiserrata and other 26 species of Camellia downloaded from NCBI GenBank using the GTR + I+G substitution model in MrBayes v3.2.6 (Fredrik et al. Citation2012), and designating Symplocos ovatilobata and Symplocos costaricana as the outgroup. The result showed that C. semiserrata, C. reticulata, C. mairei, and C. pitardii belong to the subsect. Reticulata clustered together, while C. japonica and C. chekiangoleosa belong to the subsect. Lucidissima clustered with another branch (). It showed that there are obvious differences between subsect. Reticulata and subsect. Lucidissima in sect. Camellia, which indicated that our results accord with the Flora Reipublicae Popularis Sinicae on the classification of subsections in sect. Camellia (Chang and Ren Citation1998). Besides, we also found that sect. Camellia was closely related to sect. Oleifera and sect. Paracamellia, which also reflected the flower color cannot be used as the basis for the classification of sect. Camellia (Ming Citation2000). This study provides a reference for the phylogenetic relationship of C. semiserrata and sect. Camellia species.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the GenBank accession no. MZ403753. The associated BioProject, SRA, and BioSample numbers are PRJNA750633, SRR15294181, and SAMN20475199, respectively.
Additional information
Funding
References
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120.
- Chang HT, Ren SX. 1998. Theaceae. In: Flora Reipublicae Popularis Sinicae. Vol. 49. Beijing: Science Press; p. 1–194.
- Ding XG, Zhang YZ, Chen QF, Liu YJ, Chen ZJ, Cai J, Li YQ. 2012. Law of genetic variation of fruits in Camellia semiserrata. Non-Wood Forest Res. 30(2):23–27.
- Fredrik R, Maxim T, Paul VDM, Daniel LA, Aaron D, Sebastian H, Bret L, Liang L, Marc AS, John PH. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61:539–542.
- Gu C, Ma L, Wu Z, Chen K, Wang Y. 2019. Comparative analyses of chloroplast genomes from 22 Lythraceae species: inferences for phylogenetic relationships and genome evolution within Myrtales. BMC Plant Biol. 19(1):281.
- Jin JJ, Yu WB, Yang JB, Song Y, Depamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241.
- Ming TL. 1998. The classification, differentiation and distribution of the genus Camellia sect. Camellia. Acta Bot Yunn. 20 (2):127–148.
- Ming TL. 2000. Monograph of the genus Camellia. Kunming: Yunnan Science and Technology Press; p. 293–294.
- Palmer JD. 1985. Comparative organization of chloroplast genomes. Annu Rev Genet. 19(1):325–354.
- Rong DJ, Liu J, Guo YX, Yang SW, Liang JX, Zhang YZ, Tang XX, Wang MH. 2021. Yield and fruit characters of Camellia semiserrata in Dongguan. Forest Environ Sci. 37(2):76–80.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq- versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Wu XJ, Tang L, Lin HJ, Wang YQ, Feng BM. 2008. Flavonoids from seeds of Camellia semiserrata Chi. and their estrogenic activity. Biosci Biotechnol Biochem. 72(9):2428–2431.