Abstract
Mahonia duclouxiana is a member of the genus Mahonia of Berberidaceae and is distributed in South Asia. Here, the complete chloroplast genome sequence of M. duclouxiana was reported. The complete chloroplast genome is 165,384 bp in length, which has a large single-copy (LSC) region of 73,477 bp, a small single-copy (SSC) region of 18,563 bp, and two inverted repeat (IR) regions of 36,672 bp. The G/C content in the chloroplast genome is 38.1%. The whole chloroplast genome contains 151 genes, including 38 unique tRNA genes, 105 unique protein-coding genes, and 8 unique rRNA genes. The phylogenetic analysis supported that this species should be included in Maddenia. The complete chloroplast genome sequence of M. duclouxiana will provide extremely important information in tracing the evolutionary history of the genus Mahonia and the development of the medicinal value.
Mahonia duclouxiana Gagnep. 1908, a member of the famous medicinal genus Mahonia of Berberidaceae, is distributed in the forest, shrub, roadside, or hillside of Yunnan, Sichuan, and Guangxi provinces of China. It also spread over Myanmar, India, and Thailand (Ying et al. Citation2011). Its leaves were used as traditional Chinese medicine with the effects of clearing heat, relieving cough, and resolving phlegm, and are often used to treat jaundice, colds, diarrhea, dysentery, hypertension, and bruises (Liu and He Citation2010). In this study, the complete plastid genome sequence of M. duclouxiana (GenBank accession number: MZ086770) was assembled by using the genome skimming approach (Zimmer and Wen Citation2015). The phylogenetic relationship within this genus was reconstructed.
The sample of M. duclouxiana was collected from Guiyang Medicinal Botanical Garden, Guizhou Province of China (112°43′ E, 25°46′ N, alt. 1107 m). The voucher specimen (TRCGZ005) was deposited in the Herbarium of Northwest A&F University (WUK), China. Total genomic DNA was extracted from fresh leaves by the modified cetyltrimethyl ammonium bromide (CTAB) method (Doyle and Doyle Citation1987). The libraries were prepared in the Molecular Biology Experiment Center in Southwest China using NEBNext® Ultra™ II DNA Library Prep Kit and sequenced by Illumina Miseq platform (Illumina, San Diego, CA, USA). 25,347,900 Raw Illumina data were filtered for sequence quality using Trimmomatic v. 0.40. The clean reads of the chloroplast (cp) genome were assembled by using GetOrganelle (Jin et al. Citation2020). The complete cp genome was annotated with PGA (https://github.com/quxiaojian/PGA) and corrected with DOGMA (Wyman et al. Citation2004).
The complete chloroplast genome of M. duclouxiana is 165,384 bp in length, which has the typical quadripartite structure. The length of each inverted repeat (IR) is 36,672 bp, those between them are the large single-copy (LSC) and the short single-copy (SSC) with the length of 73,477 and 18,563 bp, respectively. One hundred and fifty-one genes were found in this chloroplast genome, and 105 of them are unique protein-coding genes, 38 are unique tRNA genes and 8 are unique rRNA genes. Most of them occur a single copy, while ten tRNA genes (trnG-UCC, trnT-GGU, trnM-CAU, trnI-CAU, trnI-GAU, trnL-CAA, trnV-GAC, trnA-UGC, trnR-ACG, and trnN-GUU), 23 protein-coding genes (clpP1, psbB, psbT, pbf1, psbH, petB, petD, rps11, rpl36, infA, rps8, rpl14, rpl16, rps3, rpl22, rps19, rpl2, rpl23, ycf2, ycf15, ndhB, rps7, and ycf1) and all of the rRNA genes are duplicated owing to they are located in the IR regions. The protein-coding gene rps12 is repeated three times. The G/C content of the whole chloroplast genome length, LSC, SSC, and IR regions is 38.1%, 36.4%, 32.4%, and 41.2%, respectively.
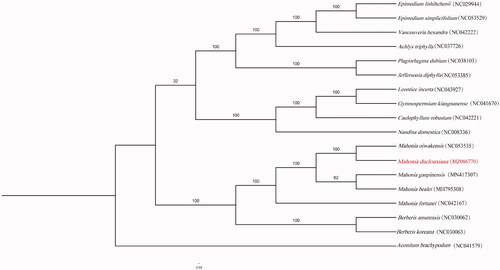
Total of 18 species (including 17 of Berberidaceae, and Aconitum brachypodum of Ranunculaceae as outgroup) were used to obtain the phylogenetic position of M. duclouxiana by RAxML with 10,000 bootstraps (). The result clearly illustrates that Mahonia duclouxiana and other species of Mahonia form a clade, which is a sister of Berberis. The complete sequence of M. duclouxiana will provide useful evidence for further phylogenetic, biogeographical, and barcoding studies.
Acknowledgments
I sincerely thank Dr. Liang Zhao for his guidance and valuable comments.
Disclosure statement
No potential conflict of interest was reported by the author.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/Genbank/update.html under the accession number MZ086770. The associated BioProject, SRA, and BioSample numbers are PRJNA727409, SRR14429474, and SAMN19020770, respectively.
Additional information
Funding
References
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Jin JJ, Yu WB, Yang JB, Song Y, de Pamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241.
- Liu AL, He SZ. 2010. Research on species and geographical distribution of Mahonia resources in China. Res Pract Mod Chinese Med. 4:20–24.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255.
- Ying JS, Boufford DE, Brach AR. 2011. Mahonia. In: Wu ZY, Raven PH, editors. Flora of China. Vol. 19. Beijing; St. Louis, MO: Science Press; Missouri Botanical Garden Press; p. 772–782.
- Zimmer EA, Wen J. 2015. Using nuclear gene data for plant phylogenetics: progress and prospects II. Next-gen approaches. J Syst Evol. 53:379–571.