Abstract
Ulva intestinalis Linnaeus 1753 (Ulvophyceae, Chlorophyta) is a marine green macroalga that is distributed on coasts of the Yellow Sea and the Bohai Sea in China. Here, the complete chloroplast genome of U. intestinalis was constructed and analyzed comparatively. The chloroplast genome of U. intestinalis is a 99,041-bp circular molecule that harbors a total of 112 genes including 71 protein-coding genes (PCGs), 26 transfer RNA genes (tRNAs), three ribosomal RNA genes (rRNAs), three free-standing open reading frames (orfs) and nine intronic orfs, and ten introns in seven genes (atpA, infA, psbB, psbC, petB, rrnL, and rrnS). The maximum likelihood (ML) phylogenomic analysis shows that U. intestinalis firstly groups with Ulva compressa, and then these two species together with the Ulva australis–Ulva fenestrata–Ulva rotundata subclade form a monophyletic clade, Ulva lineage II. U. intestinalis chloroplast genome is the only one in Ulva lineage II where the reversal of a collinear block of two genes (psbD–psbC) did not occur, and its genome structure is consistent with that of most chloroplast genomes in Ulva lineage I, indicating that the similarity of genome structure is not completely related to the genetic relationship of Ulva species. Our genomic data will facilitate the development of specific high-resolution chloroplast molecular markers for rapid identification of U. intestinalis, and help us understand its population diversity and genetic characteristics on a global scale.
The macroalgal genus Ulva Linnaeus 1753 (Ulvophyceae, Chlorophyta) includes ∼85 species of green seaweeds that grow in marine, estuarine and freshwater environments worldwide (Guiry and Guiry Citation2021). It is noticeable that Ulva species are opportunistic algae that tend to form green tides and marine fouling in nutrient-rich waters around the world (Liu et al. Citation2013; Wang et al. Citation2015). Thus far, the complete chloroplast genomes (cpDNAs) have been sequenced and constructed for 14 species in genus Ulva (e.g. Melton et al. Citation2015; Cai et al. Citation2017; Suzuki et al. Citation2018; Fort et al. Citation2021), and these Ulva cpDNAs share the same set of 100 conserved genes including 71 protein-coding genes (PCGs), 26 transfer RNA genes (tRNAs), three ribosomal RNA genes (rRNAs), and show different genome architectures due to multiple rearrangement events (Liu and Melton Citation2021).
Ulva intestinalis Linnaeus 1753 is a common marine green seaweed that has a cosmopolitan distribution with cases reported from the coasts of Asia, Australia, Europe, and America (Hayden and Waaland Citation2004; Heesch et al. Citation2009; Kirkendale et al. Citation2013; Steinhagen et al. Citation2019), and is also well-known as a typical green-tide forming macroalga (Leskinen et al. Citation2004; Kostamo et al. Citation2008). In China, this alga is distributed from temperate coasts (e.g. Yellow Sea and Bohai Sea) to subtropical coasts (e.g. South China Sea) (Titlyanova et al. Citation2012). In this study, we collected the thalli of U. intestinalis from the intertidal zone of Zhifu District (N 37°32′21″, E 121°24′29″), Yantai, Shandong Province, China in November 2020. Its specimen was deposited in the collection of marine algae in KLMEES of IOCAS (Feng Liu, [email protected]) under the voucher number 2020-YT-Uin-1. Fresh algal tissue from one individual thallus was used for DNA extraction using a Plant Genome DNA Kit (DP305, Tiangen Biotech, Beijing, China). The extracted DNA was segmented by Covaris S220 ultrasonic crater. The qualified libraries were sequenced on an Illumina NovaSeq platform (Illumina, USA) using paired-end sequencing. The complete chloroplast genome of U. intestinalis was constructed by the GetOrganelle software. The annotation of PCGs, tRNAs, rRNAs, open reading frames (orfs), and introns was performed using Open Reading Frame Finder (https://www.ncbi.nlm.nih.gov/orffinder), tRNAscan-SE 2.0 (Chan et al. Citation2021), and RNAweasel Tool (https://megasun.bch.umontreal.ca/cgi-bin/RNAweasel).
The chloroplast genome of U. intestinalis is a 99,041-bp circular molecule (GenBank accession number: MZ158703) with the G + C content of 24.97%. This genome harbored a total of 112 genes including 71 PCGs, 26 tRNAs, three rRNAs, three specific free-standing orfs, and nine intronic orfs (intron encoded proteins). Ten introns are identified to be present in seven genes including atpA (one intron), infA (one), psbB (three), psbC (one), petB (one), rrnL (two), and rrnS (one). Introns embedded in atpA, psbB, psbC, rrnL, and rrnS belong to group I intron, each of which contained an orf encoding a putative LAGLIDADG, or GIY-YIG, or H-N-H homing endonuclease. Introns embedded in infA and petB belong to group II intron (Liu and Melton Citation2021). The intron infA-62 in U. intestinalis is 596 bp in size and its intronic orf was completely degenerate. The intron petB-69 is 2216 bp in size and harbors an intronic orf which encodes a putative reverse transcriptase/maturase (RTM). The three specific orfs including orf103, orf136, and orf169 had little sequence similarity to any PCGs in the GenBank based on the search of blastp.
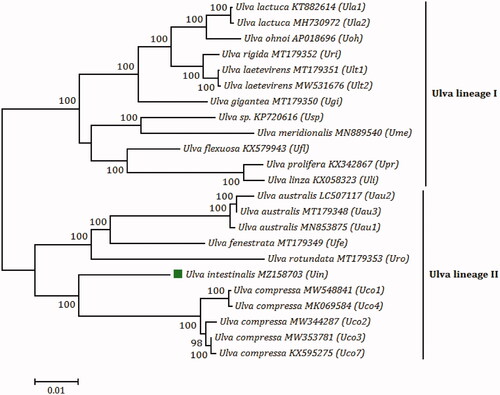
The phylogenomic tree is constructed with 1000 bootstrap replicates based on maximum likelihood (ML) analysis of amino acid (aa) sequences of 71 common PCGs from chloroplast genomes of 15 Ulva species, using MEGA 7.0. The initial tree for the heuristic search was obtained by applying the Neighbor-Joining method to a matrix of pairwise distances estimated using a JTT model (Kumar et al. Citation2016). There were a total of 22,776 positions in the final aa dataset. The phylogenomic analysis showed that 15 Ulva species fell into two clades, Ulva lineage I and II, with high bootstrap support values (100%). U. intestinalis firstly groups with Ulva compressa, and then these two species together with the Ulva australis–Ulva fenestrata–Ulva rotundata subclade forms a monophyletic clade, Ulva lineage II (). Previously, we found that a locally collinear block of two genes (psbD–psbC) has been inverted in chloroplast genomes of Ulva lineage II including U. compressa, U. australis, U. fenestrata, and U. rotundata (Liu and Melton Citation2021), whereas this reversal event did not occur in U. intestinalis cpDNA. The genome structure of U. intestinalis cpDNA is consistent with that of most chloroplast genomes in Ulva lineage I, excluding Ulva sp. and Ulva meridionalis, indicating that the similarity of genome structure is not completely related to the genetic relationship of Ulva species.
Figure 1. The maximum likelihood (ML) phylogenomic tree was constructed with 1000 bootstrap replicates based on amino acid (aa) sequences of 71 common PCGs from chloroplast genomes of 15 Ulva species, using MEGA 7.0. U. compressa MK069585 (Uco5) and U. compressa MT916929 (Uco6) were absent in this tree, due to their incomplete chloroplast genomes and faulty assembly of some PCGs (rpoA, rpoB, rpoC1, and rpoC2) (Liu and Melton Citation2021). Branch lengths are proportional to the amount of sequence change, which are indicated by the scale bar below the trees.
U. intestinalis displays great intraspecific morphological and cytological variation at different growth stages or under different environmental conditions (Leskinen et al. Citation2004), which makes it difficult to identify species solely based on morphological characteristics. Our genomic data in this study will facilitate the development of specific high-resolution chloroplast molecular markers for rapid identification of the green-tide forming alga, U. intestinalis, and help us understand its population diversity and genetic characteristics on a global scale.
Acknowledgments
We are thankful to Long Wang for the algal collection, and all the staff of the marine ecological environment genomics research group for genome assembly.
Disclosure statement
The authors are responsible for the content and writing of the paper. No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI under the accession number MZ158703 (https://www.ncbi.nlm.nih.gov/nuccore/MZ158703.1). The associated BioProject, SRA, and Bio-Sample numbers are PRJNA728799 (https://www.ncbi.nlm.nih.gov/bioproject/728799), SRR14493638 (https://trace.ncbi.nlm.nih.gov/Traces/sra/?run=SRR14493638), and SAMN19103542 (https://www.ncbi.nlm.nih.gov/biosample/SAMN19103542), respectively.
Additional information
Funding
References
- Cai C, Wang L, Zhou L, He P, Jiao B. 2017. Complete chloroplast genome of green tide algae Ulva flexuosa (Ulvophyceae, Chlorophyta) with comparative analysis. PLOS One. 12(9):e0184196.
- Chan PP, Lin BY, Mak AJ, Lowe TM. 2021. tRNAscan-SE 2.0: improved detection and functional classification of transfer RNA genes. Nucl Acids Res. gkab688. https://doi.org/10.1093/nar/gkab688.
- Fort A, McHale M, Cascella K, Potin P, Usadel B, Guiry MD, Sulpice R. 2021. Foliose Ulva species show considerable inter-specific genetic diversity, low intra-specific genetic variation, and the rare occurrence of inter-specific hybrids in the wild. J Phycol. 57(1):219–233.
- Guiry MD, Guiry GM. 2021. AlgaeBase. World-Wide Electronic Publication. Galway: National University of Ireland [accessed 2021 Aug 24]. https://www.algaebase.org.
- Hayden HS, Waaland JR. 2004. A molecular systematic study of Ulva (Ulvaceae, Ulvales) from the northeast Pacific. Phycologia. 43(4):364–382.
- Heesch S, Broom JES, Neill KF, Farr TJ, Dalen JL, Nelson WA. 2009. Ulva, Umbraulva and Gemina: genetic survey of New Zealand taxa reveals diversity and introduced species. Eur J Phycol. 44(2):143–154.
- Kirkendale L, Saunders GW, Winberg P. 2013. A molecular survey of Ulva (Chlorophyta) in temperate Australia reveals enhanced levels of cosmopolitanism. J Phycol. 49(1):69–81.
- Kostamo K, Blomster J, Korpelainen H, Kelly J, Maggs CA, Mineur F. 2008. New microsatellite markers for Ulva intestinalis (Chlorophyta) and the transferability of markers across species Ulvaceae. Phycologia. 47(6):580–587.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Leskinen E, Alström-Rapaport C, Pamilo P. 2004. Phylogeographical structure, distribution and genetic variation of the green algae Ulva intestinalis and U. compressa (Chlorophyta) in the Baltic Sea area. Mol Ecol. 13(8):2257–2265.
- Liu F, Melton JT. 2021. Chloroplast genomes of the green-tide forming alga Ulva compressa: comparative chloroplast genomics in the genus Ulva (Ulvophyceae, Chlorophyta). Front Mar Sci. 8:668542.
- Liu F, Pang S, Chopin T, Gao S, Shan T, Zhao X, Li J. 2013. Understanding the recurrent large-scale green tide in the Yellow Sea: temporal and spatial correlations between multiple geographical, aquacultural and biological factors. Mar Environ Res. 83:38–47.
- Melton JT, Leliaert F, Tronholm A, Lopez-Bautista JM. 2015. The complete chloroplast and mitochondrial genomes of the green macroalga Ulva sp. UNA00071828 (Ulvophyceae, Chlorophyta). PLOS One. 10(4):e0121020.
- Steinhagen S, Karez R, Weinberger F. 2019. Surveying seaweeds from the Ulvales and Fucales in the world's most frequently used artificial waterway, the Kiel Canal. Bot Mar. 62(1):51–61.
- Suzuki S, Yamaguchi H, Hiraoka M, Kawachi M. 2018. Mitochondrial and chloroplast genome sequences of Ulva ohnoi, a green-tide-forming macroalga in the Southern coastal regions of Japan. Mitochondrial DNA B Resour. 3(2):765–767.
- Titlyanova TV, Titlyanov EA, Xia B, Bartsch I. 2012. New records of benthic marine green algae (Chlorophyta) for the island of Hainan (China). Nova Hedwigia. 94(3–4):441–470.
- Wang Z, Xiao J, Fan S, Li Y, Liu X, Liu D. 2015. Who made the world’s largest green tide in China?—an integrated study on the initiation and early development of the green tide in Yellow Sea. Limnol Oceanogr. 60(4):1105–1117.
