Abstract
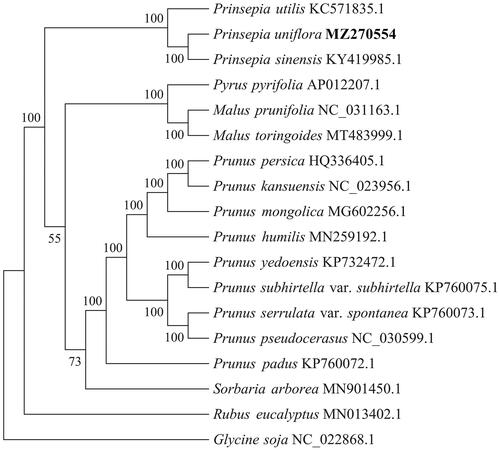
Prinsepia uniflora Batalin 1892 is a medicinal plant widely distributed in northwest China. In this study, we report and characterize the complete chloroplast (cp) genome sequence of P. uniflora. The entire sequence is 159,179 bp in length, consisting of the large single-copy region (LSC) and small single copy region (SSC) (87,239 and 19,180 bp, respectively); these two regions are separated by a pair of 26,380-bp inverted repeat (IR) regions. The genome contains 131 genes, including 86 protein-coding genes, 37 tRNA genes, and eight rRNA genes. The overall GC content of the genome is 36.7%. A phylogenetic tree constructed from 18 chloroplast genomes revealed that P. uniflora was clustered with Prinsepia sinensis and Prinsepia utilis, all of which belong to the genus Prinsepia, which is supported as a sister group by a moderate bootstrap support value of 55% with the Malus and Pyrus genera.
Prinsepia uniflora—a deciduous shrub—belongs to the genus Prinsepia (Rosaceae). It is a classic traditional Chinese medicinal plant (Zhang Citation2011; Li et al. Citation2012; Liu et al. Citation2021) and is mainly distributed in northwest China (Yang et al. Citation2008; Wang et al. Citation2012). The kernel of this plant, which is called ‘ruiren’ in traditional Chinese medicine, has long been used for the treatment of eye diseases (Zhou et al. Citation2013; Ma et al. Citation2020). Despite the importance of this plant, there is little information on the genome of P. uniflora. Here, we characterized the complete chloroplast (cp) genome sequence of P. uniflora based on genome skimming sequencing data to provide a valuable complete cp genomic resource.
Total genomic DNA was extracted from fresh leaves of P. uniflora grown in Yinchuan Botanical Garden (Yinchuan, Ningxia, China, 38°25′ N, 106°10′ E) using a modified cetyltrimethylammonium bromide (CTAB) method (Doyle and Doyle Citation1987). A specimen was deposited at the Herbarium of Yinchuan Botanical Garden (www.nxaas.com.cn; contact person: Zhu Qiang, email: [email protected]) under the voucher number RH1025; DNA (NFI-20210517RH01) was kept at the State Key Laboratory of Seeding Bioengineering, Ningxia Forestry Institute. Whole-genome sequencing was conducted using the Novaseq 6000 platform (Illumina, USA) (Genepioneer Biotechnologies Co. Ltd., Nanjing, Jiangsu, China). The filtered sequences were assembled using SPAdes assembler 3.10.1 (Bankevich et al. Citation2012). Annotation was performed using DOGMA (Wyman et al. Citation2004) and BLAST searches. A physical map was generated using OGDRAW (Greiner et al. Citation2019). A phylogenetic tree was inferred based on the maximum likelihood using RAxML v8.2.10 (Stamatakis Citation2014).
The whole cp genome of P. uniflora (GenBank accession number: MZ270554.) is 159,179 bp in length, consisting of two inverted repeats (IRa and IRb) regions of 26,380 bp, a large single copy (LSC) region of 87,239 bp, and a small single-copy (SSC) region of 19,180 bp. The cp sequence contains 131 complete genes, including 86 protein-coding genes, 37 tRNA genes, and eight rRNA genes. Intron-exon structure analysis indicated that the majority (108) of genes have no introns, whereas 19 genes have a single intron and four protein-coding genes contain two introns. The overall GC content of the cp genome is 36.65%, while the corresponding values in the LSC, SSC, and IR regions are 34.44, 30.16, and 42.66%, respectively.
To further clarify the phylogenetic characteristics of P. uniflora, alignment was performed on the 18 complete chloroplast genome sequences (17 Rosaceae chloroplast genomes and 1 Glycine soja genome as outgroup) using MAFFT v7.427 (Katoh and Standley Citation2013). Phylogenetic analysis () showed that P. uniflora was first clustered with P. sinensis and then with P. utilis, all of which belonged to the genus Prinsepia. Genus Prinsepia is supported as a sister group with the Malus and Pyrus genera with a moderate bootstrap support value of 55%. Nine species of the genus Prunus clustered together. The species Sorbaria arborea clustered together with the genus Prunus (bootstrap value 73%).Rubus eucalyptus is a separated clade. The complete cp genome sequence of P. uniflora provides useful information for conservation genetics and phylogenetic studies of this species.
Figure 1. Phylogenetic tree inferred using maximum-likelihood (ML) method based on the complete chloroplast genome of 18 representative species, including 17 Rosaceae chloroplast genomes and 1 Glycine soja genome as the outgroup. Bootstrap values are shown as percentages at the branches. The complete chloroplast genome is downloaded from the NCBI database and shown after each species name.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession no. MZ270554. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA757175, SRR15573078, and SAMN20938800, respectively.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Greiner S, Lehwark P, Bock R. 2019. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47(W1):W59–W64.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Li H, Li SY, Wang YL, Li K, Liu YQ. 2012. Physicochemical properties and fatty acid composition of Nux prinsepiae uniflorae oil. J Chinese Inst Food Sci Technol. 12(10):208–212.
- Liu JQ, Fu MJ, Wu HY, Tian B. 2021. Phylogeography of the Loess Plateau endemic plant Prinsepia uniflora inferred from the chloroplast DNA. Guihaia. 41(3):396–403.
- Ma YX, Ji JJ, Chayanis S, Gao JP, Cui YJ. 2020. Pharmacognostical study of Chinese medicine Prinsepiae nux. Chinese Tradit Herb Drugs. 51(3):763–768.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Wang YL, Li YH, Wang Y, Zhu Q. 2012. Study on seed biological characteristics and analysis of fruits nutritional components of Prinsepia uniflora. Northern Hortic. (12):53–55.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255.
- Yang FH, Zhao XM, Zhao HY, You YX. 2008. Research progress in Prinsepia uniflora. J Shanxi Agric Sci. 36(9):94–96.
- Zhang XL. 2011. The study on the ecological characteristics and development using of Prinsepia uniflora Balal. Shanxi Fores Sci Technol. 40(3):38–39.
- Zhou HY, Zhao RH, Yang JS. 2013. Two new alkaloid galactosides from the kernel of Prinsepia uniflora. Nat Prod Res. 27(8):687–690.
