Abstract
The complete chloroplast genome of Lonicera similis Hemsl. has been characterized by reference-based assembly using Illumina paired-end data. The circular complete cp genome is 155,463 bp in length, containing a large single copy (LSC) region of 89,282 bp, a small single copy (SSC) region of 18,661 bp, which are separated by a pair of inverted repeat (IR) regions of 23,760 bp. A total of 129 genes were predicted from the cp genome, including 83 protein-coding genes, 37 tRNA genes, and eight rRNA genes. Phylogenetic analysis reveals that L. similis is more closely related to Lonicera japonica Thunb. and Lonicera dasystyla Rehd. Our result will provide a reference for the phylogenetic relationship, plant identification and resource development and utilization of Lonicera species.
Lonicera similis is an important traditional Chinese medicine (TCM) plant that belongs to the Caprifoliaceae family. The buds of L. similis are used as a substitute for ‘jin yin hua’ (Lonicera japonica, a well-known TCM) in the southwest of China and have been used to treat fever, influenza, dysentery in traditional Chinese and Japanese medicine for many years (Xu et al. Citation1988; Ko et al. Citation2006). Lonicera has abundant variations in morphology and medicinal ingredients. Modern pharmacological studies show that L. similis has a higher yield of chlorogenic acid and better antibacterial activity than L. japonica (Zhang et al. Citation2015, Citation2017), however, the genetic relationship between Lonicera species is not clear, e.g. L. similis existed as an independent species of the sect. Nintooa in the Chinese version flora of China (Xu et al.Citation1988). In the updated English version flora of China (Yang et al. Citation2011), Lonicera similis and the other five species were used as synonyms for L. macrantha species complex. which seriously hindered the development and application of the Lonicera species. Chloroplast genome has been widely used to study the genetic diversity and phylogenetic relationship of plant species in recent years (Jiang Citation2019), our study will examine the phylogenetic position of L. similis in Lonicera Genus.
The leaves of L. similisis were sampled from Mt. Jingfo, NanchuanCounty, Chongqing City (28.95°N, 107.21°E), Southwest of China. The voucher specimen was deposited at Guangxi Botanical Garden of Medicinal Plants (http://www.gxyyzwy.com/, XM Wei, [email protected], GXMG: CQ00964). Total genomic DNA was extracted according to the modified CTAB method (Doyle and Doyle Citation1987). Purified genomic DNA was sequenced using the Illumina Hiseq 2500 Platform at Genepioneer Biotechnologies Inc (Nanjing, China). In total, about 24.1 million high-quality clean reads (150 bp paired-end read length) were generated. Quality trimmed were trimmed and assembled into contigs using the program SPAdes v3.10.1 (Bankevich et al. Citation2012) with the default parameters sets. The annotation was obtained after a BLAST (NCBI BLAST v. 2.2.31) search using the chloroplast genome sequence of L. japonica (GenBank accession: MH028738) as a reference sequence. Then for CDS, the annotation results were modified using prodigal v2.6.3 (Hyatt et al. Citation2010), for rRNA using hmmer v3.1b2 (Potter et al. Citation2018), and for tRNA using aragorn v1.2.38 (Dean and Bjorn Citation2004). The finally annotated chloroplast genome of L. similis has been deposited into GenBank with the accession number (MW970104).
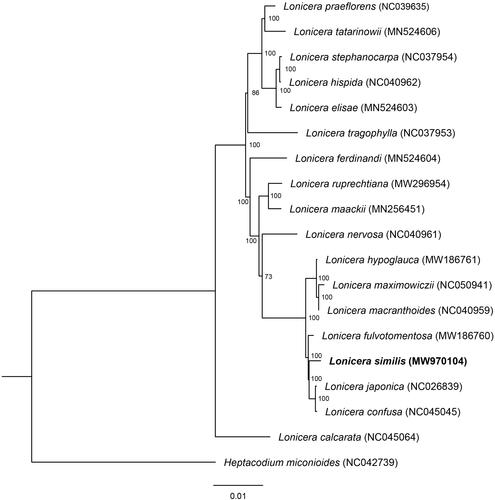
To reveal the phylogenetic position of L. similis, a phylogenetic analysis was performed based on 18 complete chloroplast genomes of Lonicera genus. Heptacodium miconioides (Genebank accession: NC042739) were chosen as outgroup. These sequences were aligned using the MAFFT v. 7.310 (Katoh and Standley Citation2013) using the published plastid genomes. The maximum-likelihood (ML) tree was constructed using RAxML v.8.2.10 (Stamatakis Citation2006) with GTRGAMMA as the nucleotide substitution model and 1000 bootstrap replicates.
The complete assembled chloroplast genome of L. similis was 155,463 bp in length. The genome presents a typical quadripartite structure with two IR (each 23,760 bp in length), separated by one SSC and one LSC region (18,661 and 89,282 bp in length, respectively). The overall GC content of L. similis plastome was 38.56%. The chloroplast genome encoded a total of 129 genes, including 83 protein-coding genes, 37 transferRNA (tRNA) genes, and eight ribosomal RNA (rRNA) genes. Phylogenetic result () showed that six species of sect. Nintooa clustered together, the L. similisis was more closely related to L. japonica and L. dasystyla. However, L. calcarata belonged to sect. Nintooa was separated from all other lonicera species, located in the basal clade, not clustering with other members within the section in taxonomy. In addition, L. maximowiczii of sect. Isika was firstly clustered with L. macranthoides, which belonged to sect. Nintooa. The complete chloroplast genome of L. similisis contributes to the phylogenetic and evolutionary analysis of Lonicera species.
Disclosure statement
The authors declare that they have no competing interests.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/nuccore/MW970104) under the accession No. MW970104. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA738588, SRR14860699, and SAMN19734401 respectively.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Dean L, Bjorn C. 2004. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 32(1):11–16.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Hyatt D, Chen G-L, Locascio PF, Land ML, Larimer FW, Hauser LJ. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 11(1):119.
- Jiang L. 2019. Characterization of the complete chloroplast genome of Lonicera japonica (Caprifoliaceae), a long history herb species plant from China. Mitochondrial DNA B Resour. 4(2):3936–3937.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Ko HC, Wei BL, Chiou WF. 2006. The effect of medicinal plants used in Chinese folk medicine on RANT ES secretion by virus-infected human epithelial cells. J Ethnopharmacol. 107(2):205–210.
- Potter SC, Aurélien L, Eddy SR, et al. 2018. HMMER web server: 2018 update. Nucleic Acids Res. 46(W1):W200–W204.
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22(21):2688–2690.
- Xu BS, Hu JQ, Wang HJ. 1988. Flora of China. Vol. 72. Beijing: Science Press; p. 143–259.
- Yang QE, Landrein S, Joanna O, Borosova R. 2011. Flora of China. Vol. 19. Beijing: Science Press; p. 616–641.
- Zhang X, Wang JR, Zhou QM, et al. 2015. Study on chemical constituents of Lonicera similis. J Chin Med Mater. 38(11):2324.
- Zhang X, Zou LH, He YL, et al. 2017. Triterpenoid saponins from the buds of Lonicera similis. Nat Prod Res. 32(19):1–9.