Abstract
Clerodendrum lindleyi Decne. ex Planch. is a Chinese medicinal plant in the Lingnan region of China. In this study, the complete chloroplast genome sequence of C. lindleyi was assembled and characterized from high-throughput sequencing data. The chloroplast genome is 151,678 bp in length, consisting of a large single-copy (LSC) and a small single-copy (SSC) regions of 83,043 bp and 17,311 bp, respectively, which are separated by a pair of 25,662 bp inverted repeat (IR) regions. The overall GC content of the genome is 38.18%. The genome contains 133 genes, including 88 protein-coding, 37 tRNA, and 8 rRNA genes. A phylogenetic tree reconstructed by using 16 chloroplast genomes reveals that C. lindleyi is most closely related to C. trichotomum which together forms a group that is a sister to genus Caryopteris. The work reported here is the first complete chloroplast genome of C. lindleyi which will provide useful information to the evolutionary studies on the genus of Clerodendrum.
Clerodendrum lindleyi Decne. ex Planch. is a kind of shrub with a special smell that can grow to 1–3 m. It is widely distributed in South China. Their dry roots and stems are used in Traditional Chinese Medicine (TCM) which have for dehumidification and detumescence in the Lingnan region of China (Chen et al. Citation2015). As a traditional Chinese medicine, it is widely used for treating bronchitis, hypertension, and rheumatism. Min et al isolated and characterized 13 bioactive compounds from C. lindleyi, including betulinic acid, monoacetylartinoside, clerodenoside A (Min et al. Citation2012). Until now, such studies have focused on the medicinal and insecticidal properties of these secondary compounds. However, the phylogenetic relationships of C. lindleyi remain unexplored and there is no genomic resources existing for research in this genus. In this study, the complete chloroplast genome of C. lindleyi was determined by using high throughput sequencing technology, which will provide valuable bioinformatic data for getter understanding the sytematics of Clerodendrum genus and forms the foundation for future genetic research.
The fresh leaves of C.lindleyi were sampled from Nanjing city, Jiangsu Province, China (31°54′10.37″N, 118°55′5.12″E). Specimens were stored in the Medical Botanical Garden of China Pharmaceutical University (accession number: Qin-JCCML-07). Total genomic DNA was extracted with a modified CTAB protocol according to Doyle and Doyle (Citation1987). The whole genome sequencing was conducted by Hefei Biodata Biotechnologies Inc. (Hefei, China) on the Illumina Hiseq platform. The filtered sequences were assembled by using the program SPAdes assembler 3.10.0 using default settings (Bankevich et al. Citation2012). The annotation was performed with DOGMA (Wyman et al. Citation2004) and BLAST searches.
The chloroplast genome of C.lindleyi is 151,678 bp in length (GenBank accession no. NC_056199) and contains inverted repeat (IR) regions of 25,662 bp, respectively, separated by large single-copy (LSC) and small single-copy (SSC) regions of 83,043 bp and 17,311 bp. The overall GC content of the C.lindleyi complete chloroplast genome is 38.18%, and the corresponding values in LSC, SSC and IR regions are 36.3%, 32.1%, and 43.3%, respectively. The complete chloroplast genome was predicted to contain 133 genes, including 88 protein-coding, 37 tRNA, and 8 rRNA genes. Eight protein-coding, six tRNA, and four rRNA genes were duplicated in the IR regions. Nineteen genes contained two exons and four genes (clpP, ycf3, and two rps12) contained thee exons.
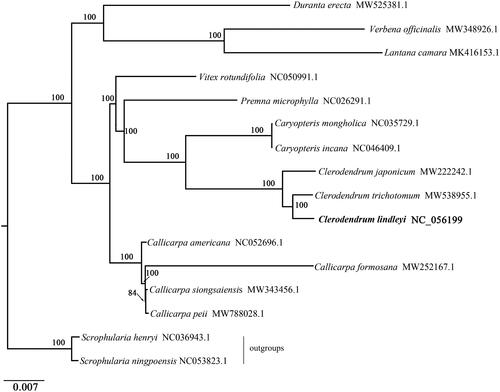
To investigate its taxonomic status, alignment was performed on the 16 complete chloroplast genome sequences using MAFFT v7.307 (Katoh and Standley Citation2013), and a Maximum Likelihood (ML) tree was constructed with 1000 bootstrap replicates in FastTree v2.1.10 (Price et al. Citation2010) with the GTR + Gamma model. Scrophularia ningpoensis and S. henryi were used as outgroups to construct the phylogenetic tree. As expected, C. lindleyi is fully resolved in a clade sister to D. trichotomum and the Clerodendrum forms a group that is a sister of the genus Caryopteris (). The complete chloroplast genome sequence of C. lindleyi will provide a useful resource for the conservation genetics of this species as well as for the phylogenetic studies of Verbenaceae.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession no. NC_056199. The associated BioProject, SRA, and Bio-sample numbers are PRJNA685332, SRS7884001 and SAMN17082918 respectively.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Chen JM, Mei QX, Peng LH, Wang HL, Cheng JL. 2015. HPLC Fingerprints of Clerodendrum lindleyi. Zhong Yao Cai. 38(6):1183–1185.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin. 19:11–15.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Min H, Zhao ZM, Guo WY, Yang DP, Cheng JL. 2012. Chemical constituents of Clerodendrum lindleyi and their free radicalscavenging activities. Chin Tradit Herb Drugs. 43(6):1050–1056.
- Price MN, Dehal PS, Arkin AP. 2010. FastTree 2-approximately maximum-likelihood trees for large alignments. PLoS One. 5(3):e9490.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255.