Abstract
Melampyrum koreanum K.-J. Kim and S.-M. Yun 2012 (Orobanchaceae) is a hemi-parasitic herb, endemic to Korea. Here, the chloroplast genome of this species is reported. It was found to be 143,865 bp long, with a large single-copy region of 83,133 bp, a small single-copy region of 10,308 bp, and a pair of inverted repeat regions of 25,212 bp each. The chloroplast genome harbors 124 genes, including 79 protein-coding genes, 37 transfer RNA genes, and eight ribosomal RNA genes. Among the identified genes, rpoA and several ndh genes were determined to be pseudogenized due to the stop codon in the middle of the gene. The phylogenetic tree of the family was reconstructed based on 20 protein-coding genes, conserved across studied taxa. As a result, Melampyrum L. 1753 species were found to form a monophyletic group in the family.
Genus Melampyrum L. 1753 of family Orobanchaceae (formerly included in family Scophulariaceae) is distributed in Eurasia and North America, and members of this genus are characterized by a hemi-parasitic lifestyle (Olmstead et al. Citation2001; Choi Citation2018). To date, four Melampyrum taxa have been recognized in Korea, but various intermediate forms among species or lower taxonomic ranks have been observed (Choi Citation2018). Such morphological features obscure taxonomic delimitation.
In Korea, an endemic species, Melampyrum koreanum K.-J. Kim and S.-M. Yun 2012, was recorded based on a longer corolla tube and style than Melampyrum roseum Maxim. 1859 (Kim and Yun Citation2012). This species grows in the islands located in the southern sea of Korea. However, it is necessary to further study this species, because of known morphological variation within/among species (Choi Citation2018). In such cases, chloroplast (cp) genome data have been utilized as an evidence for inferring the phylogenetic relationship among taxa. Here we report, for the first time, the sequence of the cp genome of M. koreanum, and compare it with the previously reported cp genome of M. roseum (Li et al., Citation2019). In addition, we analyze the phylogenetic placement of M. koreanum within Orobanchaceae.
M. koreanum was collected from Hongdo island (Hongdo-ri, Heuksan-myeon, Sinan-gun, Jeollanam-do, Republic of Korea; 34°41′3″N, 125°11′53″E). The specimen (voucher number: NIBRVP0000806190) and DNA (NIBRGR0000806190) were deposited at the National Institute of Biological Resources (KB) (https://species.nibr.go.kr/index.do; Chang Woo Hyun (specimen), [email protected]; Jinwhoa Yum (DNA), [email protected]). Genomic DNA was extracted from silica-dried leaves using a DNeasy Kit (QIAGEN, Seoul, South Korea) and paired-end (PE) libraries were prepared using a TruSeq nano DNA Kit (Illumina, San Diego, CA, USA) with a 550 bp average insert size. PE libraries were sequenced on a NovaSeq 6000 platform (Illumina). Approximately 289,653,358 of high-quality PE reads (2 × 150 bp) were generated and the reads were then mapped to the cp genome of M. roseum (GenBank: MN075942) using GetOrganelle (Jin et al. Citation2020). Genes in the cp genome of M. koreanum were annotated using GeSeq, with the M. roseum cp genome as a reference (Tillich et al. Citation2017). They were then manually edited by comparing with the genes of other genera, such as Brandisia swinglei Merr. 1918, Euphrasia regelii Wettst. 1896, Lathraea squamaria L. 1753, and Neobartsia inaequalis (Benth.) Uribe-Convers and Tank 2016, which are phylogenetically related (Bennett and Mathews Citation2006). Transfer RNAs (tRNAs) were confirmed using tRNAscan-SE (Lowe and Chan Citation2016).
Results showed that the cp genome of M. koreanum (MW463054) was 143,865 bp in length and quadripartite (large single-copy, 83,133 bp; small single-copy, 10,308 bp; a pair of inverted repeats, 25,212 bp each), with an overall GC content of 38.2%. The total number of genes in the cp genome was 124 (including 79 protein-coding genes, 37 tRNAs, and eight rRNAs), and coding regions covered 45.3% of the genome. When comparing with M. roseum, more tRNAs and pseudogenes were annotated, and duplicated ycf1 (partial gene) was excluded in M. koreanum. Six tRNA genes (trnD-GUC, trnfM-CAU, trnG-UCC, trnM-CAU, trnS-GCU, and trnS-GGA) and two pseudogenes were identified (ndhC and ndhD). In addition, some genes (rpoA, ndhB [×2], ndhJ, and ndhK) annotated in M. roseum were determined as pseudogene due to stop codon in the middle of the gene. The sequences of the pseudogenes in M. koreanum were almost completely conserved in M. roseum. We found that many ndh genes, such as ndhA, ndhF, ndhG, ndhH, and ndhI, were missing from M. koreanum. Such extensive gene loss and/or pseudogenization of photosynthetic and photorespiratory genes have previously been reported in parasitic species (Cho et al. Citation2018; Zhou et al. Citation2019).
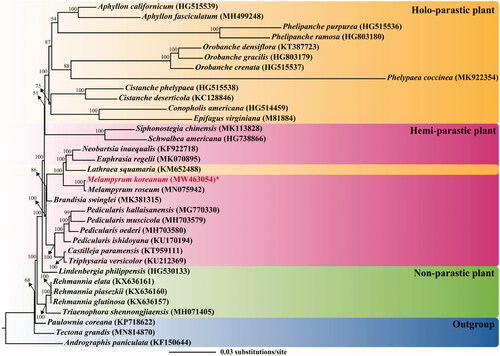
A maximum-likelihood (ML) tree of Orobanchaceae was constructed, based on 20 conserved coding regions to avoid biased topology by drastic variation among species, using IQ-TREE 1.6.12 (Trifinopoulos et al. Citation2016) with 1000 replicates (). In this analysis, 31 species from Orobanchaceae and three species from a related family were included. The TVM + I + G model was selected as the substitution model, based on the Akaike information criterion, using jModelTest 2.1.6 (Darriba et al. Citation2012). On the ML tree, all nodes were supported with strong bootstrap values (>50%), and the members of Orobanchaceae are revealed as monophyletic. In this family, it seems that members have diverged from non-parasitic to holo-parasitic life cycles, which agrees with previous research findings (Samigullin et al. Citation2016; Cho et al. Citation2018; Zhou et al. Citation2019). However, a holo-parasitic plant, L. squamaria, is nested in a clade composed of hemi-parasitic plants (including M. koreanum), implying that the transition to obligate parasitism occurred in various lineages, rather than a single-stage (Bennett and Mathews Citation2006). M. koreanum is closely related to congeneric species, M. roseum. In future studies, the inclusion of additional cp genomes of other species would enable further elucidation of the phylogenetic relationships among Korean Melampyrum taxa.
Figure 1. A maximum likelihood (ML) tree of Orobanchaceae, based on 20 coding genes of chloroplast genomes. The parasitic type of a species is represented by color rectangle: yellow, holo-parasitic plant; pink, hemiparasitic plant; green, non-parasitic plant. The number after the scientific name represents the GenBank accession of the species. The bootstrap value is shown on the node. The scale bar on the bottom indicates substitution per site. The newly sequenced individual is marked in red.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession no. MW463054. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA733910, SRR14713231, and SAMN19462170, respectively.
Additional information
Funding
References
- Bennett JR, Mathews S. 2006. Phylogeny of the parasitic plant family Orobanchaceae inferred from phytochrome A. Am J Bot. 93(7):1039–1051.
- Cho WB, Choi BH, Kim JH, Lee DH, Lee JH. 2018. Complete plastome sequencing reveals an extremely diminished SSC region in hemiparasitic Pedicularis ishidoyana (Orobanchaceae). Ann Bot Fennici. 55(1–3):171–183.
- Choi BH. 2018. Melampyrum. In: Flora of Korea, Vol. 6b. Incheon: National Institute of Biological Resources; p. 42–44.
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9(8):772–772.
- Jin J-J, Yu W-B, Yang J-B, Song Y, dePamphilis CW, Yi T-S, Li D-Z. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241 doi:https://doi.org/10.1186/s13059-020-02154-5. PMC: 32912315
- Kim KJ, Yun SM. 2012. A new species of Melampyrum (Orobanchaceae) from Southern Korea. Phytotaxa. 42(1):48–50.
- Li YC, Zhou XR, Yang J, Fu QY. 2019. Characterization of the complete chloroplast genome of Melampyrum roseum. Mitochondrial DNA B Resour. 4(2):3268–3269.
- Lowe TM, Chan PP. 2016. tRNAscan-SE on-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44(W1):W54–W57.
- Olmstead RG, de Pamphilis CW, Wolfe AD, Young ND, Elisons WJ, Reeves PA. 2001. Disintegration of the Scrophulariaeae. Am J Bot. 88(2):348–361.
- Samigullin TH, Logacheva MD, Penin AA, Vallejo-Roman CM. 2016. Complete plastid genome of the recent holoparasite Lathraea squamaria reveals earliest stages of plastome reduction in Orobanchaceae. PLOS One. 11(3):e0150718.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq–versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Trifinopoulos J, Nguyen LT, von Haeseler A, Minh BQ. 2016. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 44(W1):W232–W235.
- Zhou T, Ruhsam M, Wang J, Zhu H, Li W, Zhang X, Xu Y, Xu F, Wang X. 2019. The complete chloroplast genome of Euphrasia regelii, pseudogenization of ndh genes and the phylogenetic relationships within Orobanchaceae. Front Genet. 10:444.
