Abstract
Tabernaemontana divaricate is a vulnerable species of Apocynaceae with significant medicinal values. In this study, the complete chloroplast (cp) genome of T. divaricate was determined through the Illumina NovaSeq platform. The circular molecular genome was157,954 bp in length with two inverted repeats (IRa and IRb) regions of 25,769 bp, a large single-copy (LSC) region of 88,246 bp, and a small single-copy (SSC) region of 18,170 bp. It contained 131 genes, including 86 protein-coding, 37 tRNA, and eight rRNA genes. Phylogenetic analysis showed that T. divaricata exhibited the closest relationship with Catharanthus roseus and Rauvolfia serpentina.
The genus Tabernaemontana (Apocynaceae), comprising more than 100 species, is widespread in tropical and subtropical regions around the world (Li et al. Citation2019). Tabernaemontana divaricate (L.) R.Br. ex Roem. et Schult, also known as ‘Crape Jasmine,’ is an evergreen shrub and mainly distributed in southern China (Basavaraj et al. Citation2011). T. divaricate is an endangered species in the Redlist of China’s Biodiversity. As an important medicinal plant, T. divaricate is intensively used in antibacterial, antioxidant, analgesic, and antidiabetic treatments for its profusely high alkaloid content (Dantu et al. Citation2012; Anbukkarasi et al. Citation2016). In recent years, researches on T. divaricate have mainly focused on pharmacological properties (Naidoo et al. Citation2021), compounds identification (Li et al. Citation2019), and physiological mechanism (Thruppoyil and Ksiksi Citation2020). In this report, we characterized the complete chloroplast (cp) genome sequence of T. divaricate to contribute to further genetic and protective studies of this plant.
Fresh leaves of T. divaricata were collected from Chenshan Botanical Garden, Shanghai, China (31°08′N, 121°18′E). A voucher specimen (CSSH202105) was deposited in Shanxi Datong University (http://www.sxdtdx.edu.cn/, Kun Zhang, [email protected]). Genomic DNA was extracted according to the modified CTAB method (Doyle and Doyle Citation1987). After DNA purification, we constructed the libraries with an average length of 350 bp using the NexteraXT DNA Library Preparation Kit (Illumina, San Diego, CA, USA). High-throughput sequencing was performed on Illumina Novaseq 6000 platform, and the average length of the generated reads was 150 bp. Then 3.4 Gb clean reads were generated by editing raw sequence reads with NGS QC Tool kit (Patel and Jain Citation2012). The filtered reads were assembled by SPAdes v.3.11.0 software (Bankevich et al. Citation2012), and then the sequence of T. divaricate was annotated using PGA (Qu et al. Citation2019) with that of Rauvolfia serpentina (L.) Benth. ex Kurz (Accession number: NC_047244) as the initial reference. The annotated sequence has been submitted to NCBI (https://www.ncbi.nlm.nih.gov/nuccore/MZ073339.1/), the accession number is MZ073339.
The complete cp genome of T. divaricate was 157,954 bp in length and presented a quadripartite structure. It comprised two inverted repeats (IRa and IRb) regions, each of 25,769 bp, an 88,246 bp large single-copy (LSC) region, and an 18,170 bp small single-copy (SSC) region. The overall GC content detected in the T. divaricata cp genome was 38.1%. The assembled genome encoded 131 genes, including 86 protein-coding, 37 tRNA, and eight rRNA genes. Among the annotated genes, 15 genes (atpF, ndhA, ndhB, petB, petD, rpl2, rpl16, rpoC1, rps16, trnA-UGC, trnG-UCC, trnI-GAU, trnK-UUU, trnL-UAA, and trnV-UAC) contained one intron and two genes (clpP and ycf3) contained double introns. All genes occurred as a single copy, except that 10 genes (ycf1, rrn4.5, rrn5, rrn16, rrn23, tRNA-Ile, tRNA-Ala, tRNA-Arg, tRNA-Asn, and ORF302) were duplicated in IR regions.
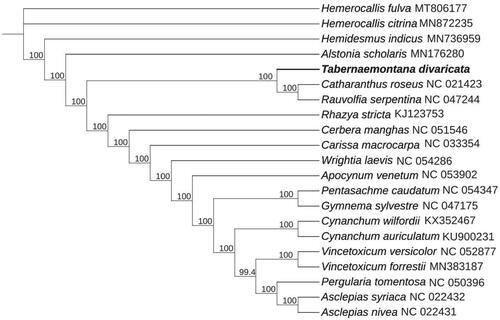
To further reveal the phylogenetic position of T. divaricata, a phylogenetic analysis was conducted with nine complete cp genomes of Apocynaceae, 12 complete cp genomes within Asclepiadaceae family, and two species [Hemerocallis citrina Baroni, Hemerocallis fulva (L.) L.] from Asphodelaceae as outgroup. The sequences were downloaded from NCBI GenBank database and aligned using MAFFT (Katoh and Standley Citation2013). A maximum-likelihood phylogenetic tree was established by IQTREE v1.6 (Jana et al. Citation2016), which indicated that T. divaricata was sister to Catharanthus roseus (L.) G. Don and Rauvolfia serpentina (). The information derived from this study provides a reference for future genetic and evolutionary surveys in T. divaricata, which may help facilitate the classification and conservation of this valuable and endangered plant.
Figure 1. Maximum-likelihood phylogenetic tree for T. divaricata based on nine complete cp genomes of Apocynaceae, 12 complete cp genomes within Asclepiadaceae family, and two species (Hemerocallis citrina and Hemerocallis fulva) from Asphodelaceae as outgroup. The bootstrap values are located on each node and the Genbank accession numbers are shown beside the Latin name of the species.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Data availability statement
The assembled complete cp genome sequence of T. divaricata has been submitted to GenBank of NCBI and is openly available under the accession number: MZ073339 (https://www.ncbi.nlm.nih.gov/nuccore/MZ073339.1/). The associated BioProject, SRA, and Bio-Sample numbers are PRJNA726627, SRR14373633, and SAMN18951226, respectively.
Additional information
Funding
References
- Anbukkarasi M, Thomas PA, Sundararajan M, Geraldine P. 2016. Gas chromatography-mass spectrometry analysis and in vitro antioxidant activity of the ethanolic extract of the leaves of Tabernaemontana divaricate. Pharmacog J. 8(5):451–458.
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Basavaraj P, Shivakumar B, Shivakumar H. 2011. Anxiolytic activity of Tabernaemontana divaricata (Linn) R. Br. flowers extract in mice. J Pharm. 2:65–72.
- Dantu AS, Shankarguru P, Ramya DD, Vedha HB. 2012. Evaluation of in vitro anticancer activity of hydroalcoholic extract of Tabernaemontana divaricate. Asian J Pharm Clin Res. 5(3):59–61.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Jana T, Lam-Tung N, Arndt VH, Quang MB. 2016. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 44:W232–W235.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Li XM, Jiang XJ, Wei GZ, Ren LH, Wang LX, Cheng XL, Wang F. 2019. New iboga-type indole alkaloids from Tabernaemontana divaricata. Nat Prod Bioprospect. 9(6):425–429.
- Naidoo CM, Naidoo Y, Dewir YH, Murthy HN, El-Hendawy S, Al-Suhaibani N. 2021. Major bioactive alkaloids and biological activities of Tabernaemontana species (Apocynaceae). Plants. 10(2):313–340.
- Patel RK, Jain M. 2012. NGS QC toolkit: a toolkit for quality control of next generation sequencing data. PLOS One. 7(2):e30619.
- Qu XJ, Moore MJ, Li DZ, Yi TS. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15(1):50–61.
- Thruppoyil SB, Ksiksi T. 2020. Time-dependent stomatal conductance and growth responses of Tabernaemontana divaricata to short-term elevated CO2 and water stress at higher than optimal growing temperature. Curr Plant Biol. 22:100127.
