Abstract
Elaeocarpus decipiens F. B. Forbes and Hemsl is an evergreen tree native to East Asia. Here, we sequenced and reported the complete chloroplast genome of E. decipiens for the first time. In the present work, the complete chloroplast (cp) genome sequence of E. decipiens was characterized by Illumina pair-end sequencing. The genome was 158,148 bp in total length, including a large single-copy (LSC) region of 85,702 bp, a small single-copy (SSC) region of 17,672 bp, and a pair of invert repeats (IR) regions of 27,387 bp. The plastid genome contained 142 genes including 97 protein-coding genes, 37 tRNA genes, and eight rRNA genes. Phylogenetic analysis based on 13 chloroplast genomes indicates that E. decipiens is closely related to Elaeocarpus bracean in Elaeocarpacea.
Elaeocarpus decipiens F. B. Forbes and Hemsl (http://powo.science.kew.org) is a broad-leaved evergreen large shrub tree native to temperate East Asia, typically found in evergreen forests from 1300 to nearly 8000 feet in elevation through much of China and in Vietnam. It typically grows from 20 to 35 feet in height with an equal spread. Its stylish branching pattern, opulent growth, solid form, and beautiful tropical foliage make this plant species an attractive landscape tree (Moore et al. Citation2020). The chloroplasts (cp) genome has a maternal inheritance and conserved structure, that has been used to examine the developmental and phylogenetic relationships of plants (Wang et al. Citation2018). For this reason, we first assembled the complete cp genome of E. decipiens based on Illumina pair-end sequencing data, to understand the phylogenetic position of the species for future evolutionary studies.
E. decipiens leaves specimens were taken from Henan Province, China (Xinyang Agriculture and Forestry University: 114°13′E, 32°17′N) and quickly dried with silica gel for DNA extraction. DNA was sent to Wuhan Bena Biotechnology Co., Ltd. to construct a DNA library and sequenced by Illumina NovaSeq 6000 sequencing platform (Illumina, San Diego, CA, USA). Fresh leaves of the individuals were collected and flash-frozen in liquid nitrogen and then stored in a refrigerator (–80 °C) until DNA extraction. The Specimens have been stored in the Herbarium of the College of Horticulture, Plant Biotechnology Laboratory, Xinyang Agriculture and Forestry University (Resource person: Guangbo Zhang and Email: [email protected]) the specimen code ED. In addition, the Illumina High-throughput sequencing platform data were filtered by the script in the NOVOPlasty (Dierckxsens et al. Citation2017). After the DNA extraction from fresh leaf tissues, its quantification was validated using Agarose gel electrophoresis and Nanodrop concentration 500 bp randomly interrupted by the Covaris ultrasonic breaker for library construction. Approximately, 14.2 GB of raw data were generated with 150 bp paired-end read lengths. The complete plastid genome of Elaeocarpus braceanus (GeneBank accession: NC_054266) as a reference and plastid genome of E. decipiens both were assembled by GetOrganelle pipe-line (https://github.com/Kinggerm/GetOrganelle), it can get the plastid-like reads, and the reads were viewed and edited by Bandage (Wick et al. Citation2015). The cp genome annotation based on the comparison was assembled by Geneious v 11.1.5 (Biomatters Ltd., Auckland, New Zealand) (Kearse et al. Citation2012).
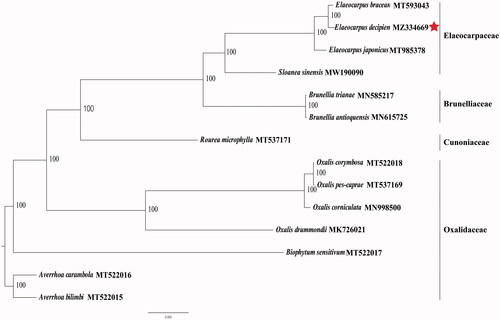
The final complete cp genome sequence of E. decipiens has been submitted to GenBank under the accession number; MZ334669. Raw reads were deposited in the GenBank Sequence Read Archive (SRA PRJNA731248). The complete cp genome of E. decipiens was a circular shape of 158,148 bp in length, consisting of four distinct regions: a large single-copy (LSC), a small single-copy (SSC), and a pair of inverted repeats (IR) with regions of 85,702 bp, 17,672 bp, and 27,387 bp, respectively. The complete cp genome consisted of 142 genes, including 97 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. The complete cp genome GC content was 34.32%. Phylogenetic analyses including E. decipiens, E. bracean, Elaeocarpus japonicus, Sloanea sinensis, seven Oxalidaceae species, two Brunelliaceae species, and one Cunoniaceae species, were performed to use complete cp genomes. All of them were downloaded from NCBI GenBank. The sequences were aligned by MAFFT v7.307 (Katoh and Standley Citation2013), and the phylogenetic tree was constructed by RAxML (Stamatakis Citation2014). Both the E. decipiens and E. bracean, belonging to the Elaeocarpacea family, had a close relationship. The analysis of the cp genome of E. decipiens provides excellent genetic information for further studies of this precious species and the taxonomy, phylogenetics, and evolution of Elaeocarpacea ().
Acknowledgments
The author wishes to thank the anonymous reviewers who provided constructive comments and critical insight on this article.
Disclosure statement
The authors reported no possible conflicts of interest.
Data availability statement
The data that support the findings of this study are openly available in GeneBank at https://www.ncbi.nlm.nih.gov/. The complete chloroplast genome generated for this study has been deposited in GeneBank with accession number MZ334669. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA731248, SRX10944172, and SAMN19276134, respectively.
Additional information
Funding
References
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Moore MR, Brito JA, Qiu S, Roberts CG, Combee LA. 2020. First report of Meloidogyne enterolobii infecting Japanese blue berrytree (Elaeocarpus decipiens) in Florida, USA. J Nematol. 52(5):1–3.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Wang J, Li C, Yan C, Zhao X, Shan S. 2018. A comparative analysis of the complete chloroplast genome sequences of four peanut botanical varieties. PeerJ. 6:e5349.
- Wick RR, Schultz MB, Zobel J, Holt KE. 2015. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics. 31(20):3350–3352.