Abstract
Artocarpus champeden Spreng. is a popular fruit tree, grown in tropical and subtropical regions. Besides food, A. champeden is also a medicinal plant with various medicinal properties. In this study, A. champeden chloroplast genome was sequenced, assembled, and annotated due to its rich information on species evolution and inter-species genetic relationships. The quadripartite structure of A. champeden complete chloroplast genome is 158,568 bp in length and comprises a large single-copy region (LSC) of 88,076 bp, a small single-copy region (SSC) of 19,028 bp, and two inverted repeat regions (IRa and IRb) of 25,732 bp. A total of 131 genes were annotated, including 85 protein-coding genes, 37 tRNA genes, eight rRNA genes, and one pseudogene. Phylogenetic analysis revealed a close relationship between A. champeden and A. heterophyllus. In addition, the study provides abundant genomic information for future phylogenetic studies of A. champeden and the Moraceae family.
Artocarpus champeden (Lour.) Spreng. (A. integer (Thunb.) Merr. 1917) is a popular fruit tree belonging to the family Moraceae (Widyawaruyanti et al. Citation2007). It is traditionally grown in tropical and subtropical regions, particularly in Southeast Asian countries, including Thailand, Peninsular Malaysia, Myanmar, Vietnam, and Indonesia (Syah et al. Citation2006; Lim et al. Citation2011). Both the flesh and seeds of A. champeden fruits are edible. The seeds are a good source of dietary fiber and resistant starch (Zabidi and Aziz Citation2009). Moreover, the wood of A. champeden is used commercially as timber (Achmad et al. Citation1996).
Artocarpus champeden is also a medicinal plant with various medicinal properties, including antimalarial, anticancer, and cell adhesion activities (Lopes et al. Citation2018). The seeds contain lectins such as IgA1-reactive and D-galactose-binding lectin. These lectins are used in biomedical research to detect tumors and identify glycoproteins (Lim et al. Citation1997). The wood pulp of A. champeden has essential bioactive compounds, including ascorbic acid and total phenolics (Lopes et al. Citation2018). Several isoprenyl flavonoids with cytotoxicity (Hakim et al. Citation1999) and antimalarial activity (Boonlaksiri et al. Citation2000) have been isolated from A. champeden.
While the economic potential of A. champeden is due to its food and medicinal value, very little research on its genome exists. Chloroplast genomes are rich in genetic information and are fundamental in evolutionary studies. This is the first study to sequence, assemble, and annotate the chloroplast genome of A. champeden.
Artocarpus champeden was collected from the Xishuangbanna Tropical Flowers and Plants Garden (100.785968 E, 22.012288 N), and the genomic DNA was extracted using the DNeasy Plant Mini Kit (Qiagen). The specimen and DNA were deposited in the herbarium and cryogenic sample library of the Yunnan Institute of Tropical Crops (http://www.yitc.com.cn, Dr. Jin Liu, [email protected]) with voucher numbers YITC-2020-FZ-M-028 and D2020-FZ-M-028, respectively. Paired-end (PE) sequencing was conducted using the Illumina HiSeq 2500 platform (Illumina, San Diego, CA, USA) with 350 bp library insert sizes. The clean reads were assembled with SPAdes-3.5.0 (http://soap.genomics.org.cn/soapdenovo.html) based on sequence overlap and paired-end relationships. Kmer 79 and Kmer 97 values were used, and Sanger sequencing verified the four boundaries of the IR region. Annotation was performed by CpGAVAS2 (Shi et al. Citation2019) and ORF Finder (https://www.ncbi.nlm.nih.gov/orffinder/). The chloroplast genome data of A. champeden was submitted to GenBank (http://www.ncbi.nlm.nih.gov/) under the accession number MT900597.
The quadripartite structure of A. champeden whole chloroplast genome is 158,568 bp in length, and comprises 49,698 A bases (31.34%), 51,343 T bases (32.38%), 28,466 G bases (17.95%), and 29,061 C bases (18.33%). Moreover, it contains a large single-copy region (LSC) of 88,076 bp, a small single-copy region (SSC) of 19,028 bp, and two inverted repeat regions (IRa and IRb) of 25,732 bp. The total GC content of the whole chloroplast genome of A. champeden is 36.28%, with corresponding values of the LSC, SSC, and IR regions of 33.96%, 29.47%, and 42.76%, respectively. In total, 131 genes, including 85 protein-coding genes, 37 tRNA genes, 8 rRNA genes and 1 pseudogene, were annotated.
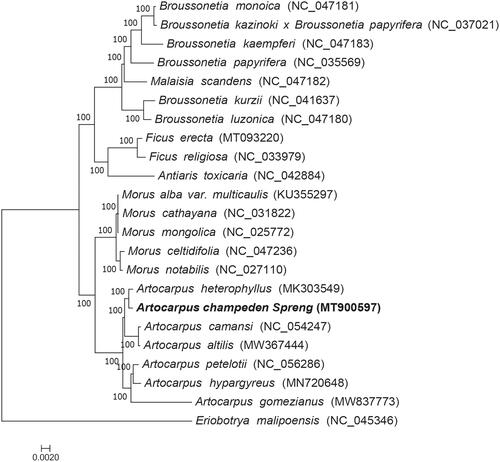
Chloroplast genome sequences of 22 species () of Moraceae family were downloaded from GenBank and used to study the phylogeny of A. champeden, Multiple sequence alignment was performed using MAFFT (Katoh and Standley Citation2013), The jModelTest 2.1.7 (David Citation2008) software was employed to analyze nucleotide substitutions model under the Akaike Information Criterion (AIC), the GTR + G + I model was selected for nucleotide and the maximum-likelihood analysis was conducted with RAxML8.2.4 (Stamatakis Citation2014). Eriobotrya malipoensis, which belongs to the Rosaceae family, was the out-group, and the node support was estimated from the results of 1000 bootstrap replicates. The phylogenetic analysis revealed a close relationship between A. champeden and A. heterophyllus (). This study provides abundant genomic information for future phylogenetic studies of the Moraceae family.
Disclosure statement
No potential competing interest was reported by the authors.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MT900597. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA703082, SRR13781753, and SAMN18011290 respectively.
Additional information
Funding
References
- Achmad SA, Hakim EH, Juliawaty LD, Makmur L., Suyatno 1996. A new prenylated flavone from Artocarpus champeden. J Nat Prod. 59(9):878–879.
- Boonlaksiri C, Oonanant W, Kongsaeree P, Kittakoop P, Tanticharoen M, Thebtaranonth Y. 2000. An antimalarial stilbene from Artocarpus integer. Phytochemistry. 54(4):415–417.
- David P. 2008. jModelTest: phylogenetic model averaging. Mol Biol Evol. 25(7):1253–1256.
- Hakim EH, Fahriyati A, Kau MS, Achmad SA, Makmur L, Ghisalberti EL, Nomura T. 1999. Artoindonesianins A and B, two new prenylated flavones from the root of Artocarpus champeden. J Nat Prod. 62(4):613–615.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Lim LBL, Chieng HI, Wimmer FL. 2011. Nutrient composition of Artocarpus champeden and its hybrid (Nanchem) in Negara Brunei Darussalam. AJSTD. 28(2):122–138.
- Lim SB, Chua CT, Hashim OH. 1997. Isolation of a mannose-binding and IgE- and IgM-reactive lectin from the seeds of Artocarpus integer. J Immunol Methods. 209(2):177–186.
- Lopes MMDA, Souza KOD, Silva EDO. 2018. Cempedak—Artocarpus champeden. Exotic Fruits. 121–127. https://doi.org/https://doi.org/10.1016/B978-0-12-803138-4.00017-4
- Shi LC, Chen HM, Jiang M, Wang LQ, Wu X, Huang LF, Liu C. 2019. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47(W1):W65–W73.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Syah YM, Juliawaty LD, Achmad SA, Hakim EH, Ghisalberti EL. 2006. Cytotoxic prenylated flavones from Artocarpus champeden. J Nat Med. 60(4):308–312.
- Widyawaruyanti A, Subehan Kalauni SK, Awale S, Nindatu M, Zaini NC, Syafruddin D, Asih PBS, Tezuka Y, Kadota S. 2007. New prenylated flavones from Artocarpus champeden, and their antimalarial activity in vitro. J Nat Med. 61(4):410–413.
- Zabidi MA, Aziz NAA. 2009. In vitro starch hydrolysis and estimated glyceamic index of bread substituted with different percentages of champedak (Artocarpus integer) seed flour. Food Chem. 117(1):64–68.