Abstract
Ammopiptanthus mongolicus is the only evergreen broadleaf shrub in the northwest desert of China, which can survive in long-term aridity and extremely cold environments. In this study, the complete chloroplast genome sequence of A. mongolicus was reported based on the Illumina NovaSeq Platform (Illumina, San Diego, CA). The chloroplast genome is 156,077 bp in length, containing a pair of inverted repeated (IR) regions (14,698 bp) that are separated by a large single copy (LSC) region of 88,025 bp, and a small single copy (SSC) region of 36,606 bp. Moreover, a total of 115 functional genes were annotated, including 81 mRNA, 30 tRNA genes, and 4 rRNA genes. Phylogenetic analysis based on 16 chloroplast genomics indicates that A. mongolicus is closely related to A. nanus.
Ammopiptanthus mongolicus is the only evergreen broadleaf shrub in the northwest desert of China, and is an endangered survivor from the Tethys in the Tertiary Period, which could grow well in long-term aridity and extremely cold environments (Liu et al. Citation2013). The habitats of A. mongolicus are marked by arid climate with an annual precipitation less than 200 mm and evaporation up to 3000 mm. Its natural distribution areas are also characterized as sandy or stony soil with high salinity, intense ultraviolet irradiation, and seasonally extreme temperature from about −30 °C in winter to more than 40 °C in summer. The extreme tolerance of this species to harsh environments makes it invaluable for exploring key stress-tolerant genes and mechanisms, especially those involved in cold and drought tolerance. Therefore, A. mongolicus has extremely important protective genetic value (Wu et al. Citation2014). However, genetic diversity of A. mongolicus has not yet been reported. Here we determined the complete sequence of the chloroplast genome of this plant compared the structures of genes and the genome of the A. mongolicus chloroplast with those of other plant species.
The fresh and young leaves of wild A. mongolicus collected from Minqin County (39°16´36.8〃N, 103°45´29.1〃E; elevation 1625 m) in Gansu Province, China. The voucher specimen and extracted DNA were deposited at Herbarium of Gansu Desert Control Research Institute under the number AM2020520-1 (Li Chang, [email protected]). Total genomic DNA extraction and genome sequence assembling were conducted by Benagen Corporation (Wuhan, China). The locations of protein-coding genes were determined by comparing with the corresponding sequences of other species.
The complete genome of A. mongolicus is 156,077 bp in length with a typical quadripartite structure, containing a pair of inverted repeated (IR) regions (14,698 bp) that are separated by a large single copy (LSC) region of 88,025 bp, and a small single copy (SSC) region of 36,606 bp. The GC content of the whole genome was 36.87%. A total of 115 functional genes were annotated, including 81 mRNA, 30 tRNA genes, and 4 rRNA genes. The protein-coding genes, tRNA genes, and rRNA genes account for 70.43%, 26.09%, and 3.48% of all annotated genes, respectively.
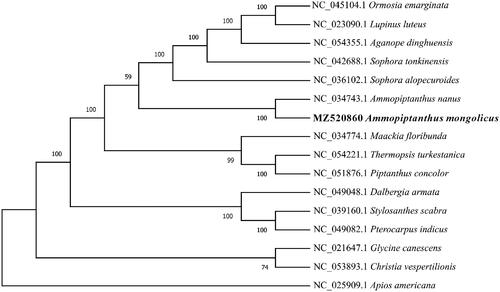
The maximum-likelihood phylogenetic tree (ML tree) was generated based on the complete genome of A. mongolicus and other species of the family Leguminosae (). Alignment was conducted using MAFFT (Katoh and Standley Citation2013), and the phylogenetic tree was built using RAxML (Stamatakis Citation2014). The results showed that A. mongolicus was closely related to A. nanus, supporting the view of A. mongolicus belongs to the genus Ammopiptanthus ().
This study provided the characterization of the complete chloroplast genome structure of A. mongolicus and showed the phylogenetic relationships in Legume. All information about A. mongolicus chloroplast genome will provide a fundamental genetic reference for future efficient breeding of new trees and ornamental woody plants with improvement in abiotic stress tolerance.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at database (https://www.ncbi.nlm.nih.gov) under the accession number MZ520860. The associated Bio-Project, SRA, and Bio-Sample numbers are PRJNA743149, SRR15039806, and SAMN19999421, respectively.
Additional information
Funding
References
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Liu M, Shi J, Lu C. 2013. Identification of stress-responsive genes in Ammopiptanthus mongolicus using ESTs generated from cold- and drought-stressed seedlings. BMC Plant Biol. 13(1):88–114.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Wu Y, We W, Pang X, Wang X, Zhang H, Dong B, Xing Y, Li X, Wang M. 2014. Comparative transcriptome profiling of a desert evergreen shrub, Ammopiptanthus mongolicus, in response to drought and cold stresses. BMC Genomics. 15(1):1–16.