Abstract
Dictyostelium intermedium is a member of dictyostelids, the unicellular eukaryotes with a unique life cycle, including a social cycle. Despite the high diversity of dictyostelids, only five species' complete mitochondrial genome sequences were reported. This study aimed to add the D. intermedium mitochondrial genome sequence to the list. The size of this genome is 58,627 bp, with 73.99% A/T, containing 62 genes located on one strand: 41 protein-coding genes, three ribosomal RNA genes, and 18 transfer RNA genes. The 41 protein-coding genes comprised 18 oxidative phosphorylation-related, 16 ribosomal, and seven hypothetical protein-coding genes. The cox1/2 and rnl gene contained introns, similar to other species of Dictyostelium. The phylogenetic tree built based on 34 protein sequences supported the monophyletic clade of Dictyostelium and the dictyostelids' ancestor's position between the two dictyostelids orders: Dictyosteliales and Acytosteliales.
Dictyostelium intermedium sp. n. is a member of dictyostelids, the highly diverse group of unicellular eukaryotes with a unique life cycle consisting of a vegetative, sexual, and social cycle (Romeralo et al. Citation2012). It was firstly isolated from the forest humus and leaf mold, Peutjang Island, Java, Indonesia, and deposited at the American Type Culture Collection (ATCC) (https://www.atcc.org, email: [email protected]) by Professor James C. Cavender (Cavender Citation1976). The nuclear gene phylogenies showed that it was clustered within the monophyletic clade of Dictyostelium (Schilde et al. Citation2019), one out of 12 recently classified genera (Sheikh et al. Citation2018). The nuclear protein-coding gene phylogeny further showed that Dictyostelium could be separated into five subclades and D. intermedieum is clustered with D. discoideum in one subclade (Schilde et al. Citation2019). Because only five species' complete mitochondrial genome (mitogenome) sequences were publicly available (Heidel and Glöckner Citation2008), this study aimed to add D. intermedium mitogenome to the list.
The 454 short-read whole-genome sequences of D. intermedium strain PJ-11 were retrieved from the ENA database under the accession SRR037009-17. These data were submitted by the Baylor College of Medicine Human Genome Sequencing Center (BCM-HGSC). The quality of short reads was evaluated by FastQC (Andrews Citation2010), then the reads were mapped to the genome of Escherichia coli (NC_000913) and Klebsiella aerogenes (NC_015663), the two bacteria commonly used for culturing dictyostelids, using BWA-MEM (Li Citation2013). The unmapped reads from SRR037010-11 and SRR037013-14 were assembled by Unicycler (Wick et al. Citation2017). The contig with the length between 40 and 60 kb was annotated. The mitochondrial protein-coding genes (PCGs) were identified by blastx (Camacho et al. Citation2009), MUSCLE (Edgar Citation2004), and ORFfinder (Souvorov et al. Citation2008). For ribosomal RNA (rRNA) genes, the small subunit rRNA gene (rns) was predicted by RNAmmer (Lagesen et al. Citation2007), while blastn (Camacho et al. Citation2009) and MUSCLE (Edgar Citation2004) were applied for identifying the intron-containing large subunit rRNA (rnl) and 5S rRNA gene. The databases for blastx and blastn were extracted from previously reported dictyostelids mitogenomes: D. discoideum (NC_000895), D. citrinum (DQ336395), Heterostelium pallidum (NC_006862), H. album (EU275726), and Cavenderia fasciculata (NC_010653). The nucleotide Ns were manually added to correct the reading frame of some protein-coding genes and then replaced by the highest frequency nucleotides found among short reads mapped to the N-containing regions in these genes by BWA-MEM (Li Citation2013). The tRNAscan-SE (Lowe and Chan Citation2016) with manual curation was applied to predict transfer RNA (tRNA) genes.
Amino acid sequences translated from mitochondrial PCGs were retrieved from six dictyostelids species and an outgroup (Acanthamoeba castellanii: NC_001637). Each gene was aligned using MUSCLE (Edgar Citation2004) and manually edited on Aliview (Larsson Citation2014), then concatenated by catfasta2phyml tool (Nylander Citation2011). The sequence matrix was applied to MEGA X (Kumar et al. Citation2018) for identifying the optimal evolutionary model. The maximum likelihood (ML) phylogeny was built by the RAxML (Silvestro and Michalak Citation2012) with 1000 bootstrap replicates.
The total length of Dictyostelium intermedium mitogenome is 58,627 bp (TPA: BK014289), comprising 45.30% A, 28.69% T, 17.02% G, and 8.99% C. This mitogenome contains 18 oxidative phosphorylation (OXPHOS) related PCGs, 16 ribosomal PCGs, seven hypothetical PCGs, three rRNA genes, and 18 tRNA genes. The cox1/2 and rnl genes contained 3 and 2 introns, respectively. These genes are located on one strand. By revising the potential function of 11 hypothetical PCGs in the previously reported dictyostelids mitogenomes by blastx, 18 OXPHOS related PCGs, 18 ribosomal PCGs, and three rRNA genes have commonly been found. The paralogs of rps3 and rps11 gene could not be identified on the D. intermedium mitogenome; however, due to the conserved position across the previously reported five species, the two mitogenome regions located next to rps3 and rps11 gene possibly carrying these paralogs. The arrangement of non-hypothetical PCGs and all rRNA genes of D. intermedium is similar to that found in D. discoideum and D. citrinum, but not D. purpureum (Heidel and Glöckner Citation2008). Therefore, the mitochondrial genome of D. intermedium has size, content, and arrangement, similar to other species of Dictyostelium.
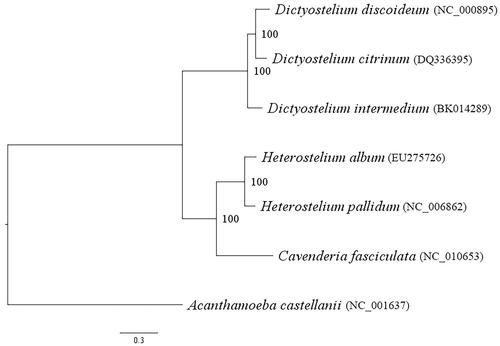
Amino acid sequences translated from 34 mitochondrial genes were retrieved from seven species, including A. castellanii. The maximum likelihood phylogeny was built based on the LG substitution model and a gamma correction for rate variation across sites with empirical frequencies (LG + G + F), as shown in . This phylogeny presented two monophyletic clades: one contained only Dictyostelium, and another contained Heterostelium and Cavanderia, which corresponded to two orders: Dictyosteliales and Acytosteliales (Sheikh et al. Citation2018). The common ancestor of dictyostelids was placed between these two clades. The topology of this phylogenetic tree corresponded well with mitochondrial genome phylogeny (Heidel and Glöckner Citation2008), 18S rRNA gene phylogeny (Sheikh et al. Citation2018), and nuclear PCGs phylogeny (Schilde et al. Citation2019). Therefore, this phylogeny supported the monophyletic group of Dictyostelium and dictyostelids' ancestor's position between Dictyosteliales and Acytosteliales.
Supplemental Material
Download MS Word (37.7 KB)Disclosure statement
No potential competing interest was reported by the author(s).
Data availability statement
The complete mitogenome sequence of Dictyostelium intermedium is available in the Third Party Annotation Section of the DDBJ/ENA/GenBank (https://www.ncbi.nlm.nih.gov) databases under the accession number TPA: BK014289. While waiting for the availability of the updated sequence record, the mapping results supporting the replacement of Ns in the sequence and the updated sequence are available upon request. This data was derived from the 454 short-read whole-genome sequence data of D. intermedium submitted by the Baylor College of Medicine Human Genome Sequencing Center (BCM-HGSC) and stored in the ENA database at https://www.ebi.ac.uk/ena/browser/home, accession number SRR037009-17.
Additional information
Funding
References
- Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data. Cambridge, United Kingdom: Babraham Bioinformatics, Babraham Institute.
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics. 10(1):421.
- Cavender JC. 1976. Cellular slime molds of Southeast Asia. I. Description of new species. Am J Bot. 63(1):60–70.
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32(5):1792–1797.
- Heidel AJ, Glöckner G. 2008. Mitochondrial genome evolution in the social amoebae. Mol Biol Evol. 25(7):1440–1450.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Lagesen K, Hallin P, Rødland EA, Staerfeldt H-H, Rognes T, Ussery DW. 2007. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 35(9):3100–3108.
- Larsson A. 2014. AliView: a fast and lightweight alignment viewer and editor for large datasets. Bioinformatics. 30(22):3276–3278.
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. ArXiv. 1303.
- Lowe TM, Chan PP. 2016. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44(W1):W54–W57.
- Nylander J. 2011. Catfasta2pyml.pl. [accessed 2014 May 15]. Available from http://www.abc.se/∼nylander/catfasta2phyml/.
- Romeralo M, Escalante R, Baldauf SL. 2012. Evolution and diversity of dictyostelid social Amoebae. Protist. 163(3):327–343.
- Schilde C, Lawal HM, Kin K, Shibano-Hayakawa I, Inouye K, Schaap P. 2019. A well supported multi gene phylogeny of 52 dictyostelia. Mol Phylogenet Evol. 134:66–73.
- Sheikh S, Thulin M, Cavender JC, Escalante R, Kawakami S-I, Lado C, Landolt JC, Nanjundiah V, Queller DC, Strassmann JE, et al. 2018. A new classification of the dictyostelids. Protist. 169(1):1–28.
- Silvestro D, Michalak I. 2012. raxmlGUI: a graphical front-end for RAxML. Org Divers Evol. 12(4):335–337.
- Souvorov A, Landsman D, Lipman DJ, Church DM, Benson DA, Maglott DR, Sequeira E, Yaschenko E, Schuler GD, Starchenko G, et al. 2008. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 36(Database issue):D13–D21.
- Wick RR, Judd LM, Gorrie CL, Holt KE. 2017. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol. 13(6):e1005595.