Abstract
In this study, the complete chloroplast (cp) genome of Ixora chinensis was sequenced by next-generation sequencing for the first time. The complete cp genome is 154,787 in length and contained 131 genes, consisting of 86 protein-coding genes, eight ribosomal RNA genes, and 37 transfer RNA genes. The phylogenetic position based on chloroplast genomes suggests that I. chinensis was closely related to I. chinensis (MN850660.1) within the Ixora clade, which may provide useful information for further understanding the evolution of I. chinensis.
Subject classification code:
Ixora chinensis Lam. also named ‘Dauk Chem’ is native to China and Malaysia, belonging to the genus of Ixora in the Rubiaceae family. In recent years, the flower has become more and more popular in China. It is widely distributed in the southwest and southeast regions of China, such as Hainan, Fujian, Guangdong and Guangxi (Li et al. Citation2019). Ixora chinensis has a longer vase life than other Ixora species which makes it an outstanding cut flower (Chen et al. Citation2003). In addition, most of the activities reported on leaves and flowers of I. chinensis were anti-oxidants, anticancer, anti-microbial activities (Bhagyasri et al. Citation2019). Interestingly, the flower with bright and abundant flowers has a long blooming term lasting for almost the entire year. Ixora chinensis with red flowers is one of the most common native species in southern China, not only with ornamental value but more importantly with its medicinal values which it is rich in flavonoids and anthocyanins (Rastogi and Mehrotra Citation1990). Recently, I. chinensis was assembled by Ly et al. (Citation2020) to comparative genomics and molecular phylogeny in subfamily Ixoroideae. However, it was not assembled into a circular chloroplast genomes and the cp genome was not annotated. In addition, the chloroplast sequence (accession number: MN850660.1) is not verified by NCBI. There is yet little information on the complete chloroplast (cp) genome of I. chinensis. Here, the complete chloroplast (cp) genome of I. chinensis was assembled and annotated for the first time. We also carried out a systematic comparative genomic study of I chinensis plastome and those from another two Ixora species, in order to explore its phylogenetic relationship and identify new regions of genomic variability.
The plant material of fresh leaves for extraction was collected from Caiban Village, Jiuhu Town, Zhangzhou (Fujian, China, 117°59′56″E, 24°48′40″N). Total chloroplast DNA was extracted following Kim et al. (Citation2006) with the optimized CTAB method, and used to construct to build an Illumina pair-end library. Then the library was done on Illumina Novaseq 6000 platform at Beijing Genomics Institute (BGI, Shenzhen, China) and generating approximately 3.6 G of raw data. The quality of reads was filtered using the FastQC program (Andrews Citation2014). High-quality clean reads were de novo assembled using SPAdes version 3.11.0 software (Bankevich et al. Citation2012) the cp genome was annotated using the online program GeSeq (Tillich et al. Citation2017) with the chloroplast genome of Vangueria infausta (accession number MN851269) as reference. The annotated chloroplast genome sequence of was deposited in GenBank under the accession number MZ221832. The voucher specimens of I. chinensis was stored at laboratory of Department of Biological Science and Biotechnology, Minnan Normal University, Zhangzhou (URL: https://bio.mnnu.edu.cn/info/1110/2223.htm, accession num-ber: No.MNU001) (Ana Bian, Email: [email protected]).
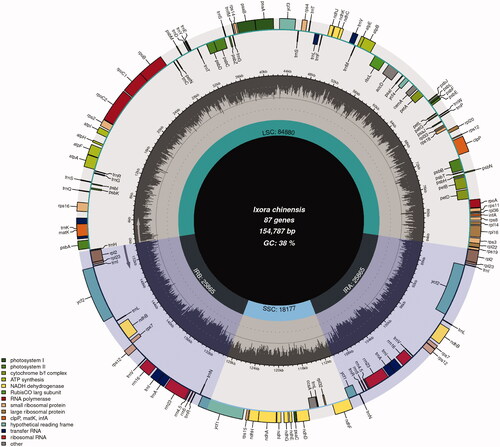
The complete length of I. chinensis is 154,787 bp (), longer than I. chinensis (MN850660.1, length = 154,665 bp, GC = 37.48%) and overall GC content of 37.5%. The cp genome of I. chinensis has a typical quadripartite structure, containing a large single-copy region of 84,880 bp, a small single-copy region of 18,177 bp, and a pair of inverted repeat regions of 25,865 bp, is longer than I. chinensis (MN850660.1, LSC = 84,874 bp, SSC= 18,157 bp, IR = 25,817 bp). We annotated the cp genome of I. chinensis (MN850660.1). The results show that there is no difference in the gene contents between I. chinensis (MZ221832.1) and I. chinensis (MN850660.1). The CP genome comprised 131 genes, including 86 protein-coding genes, 37 transfer RNA genes (tRNA), and 8 ribosomal RNA genes (rRNA). The chloroplast genome of I. chinensis has 17 different intron containing genes, exhibiting two introns (clpP and ycf3) and the rest of the genes contained one intron (trnK-UUU, rps16, trnG-UCC, atpF, rpoC1, trnL-UAA, trnV-UAC, petB, petD, rpl16, rpl2, ndhB, trnI-GAU, trnA-UGC, ndhA).
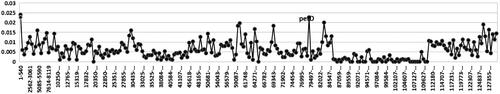
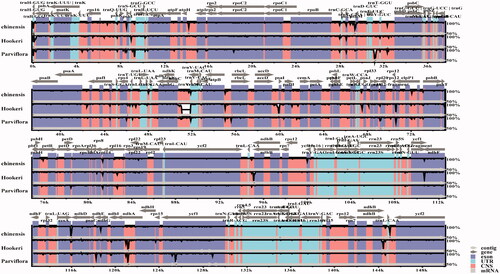
To explore the ixora species’ gene gene pseudonization and deletion scale, we performed a two-way clustering analysis of 3 Ixora species and use A. thaliana as a reference (). We found that the tRNA (trnA-Y) and genes from the ycf family (ycf5, ycf6, ycf9, ycf10) were lost in all Ixora plastomes compared with that of A. thaliana (). Polymorphism analysis of 3 Ixora species shows that petD gene has a higher variation (). Sequence divergence was visualized using mVISTA with I. chinensis (MN850660.1) as the reference annotated genome. Globally, sequence divergence among all ixora was relatively high and mainly concentrated in conserved non-coding sequences and in Untranslated Transcribed Regions (UTR). However, variation among species seemed to be negligible for UTRs located in the IR region. Substitutions were more frequent but indels (trnV-UAC) were observed as well in I. hookeri ().
Figure 4. Sequence identity plot comparing four species of subfamily Ixora with I. chinensis (MN850660.1) as the reference annotated genome.
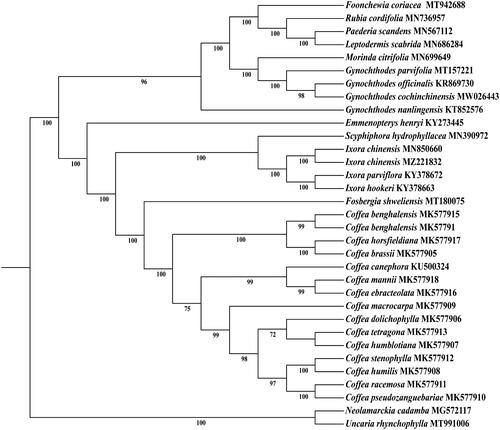
In order to understand the phylogenetic relationship between I. chinensis and other species of Ixora, the complete chloroplast sequence of 43 species was downloaded from NCBI as the phylogenetic dendrogram. The tree phylogenetic with maximum likelihood (ML) analysis method was performed using RaxML software v 8.2.9, of which the bootstrap values were calculated using 1000 replicates under the GTRGAMMAI substitution model in CIPRES (Stamatakis Citation2014). The reconstructed phylogeny showed that I. chinensis had the closest relationship with I. chinensis (MN850660.1) (), which can provide a theoretical basis for further study on the phylogeny and evolution of Rubiaceae plants ().
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference number MZ221832.
The associated BioProject, SRA, and Bio-Sample numbers and are PRJNA743157 SRR15020882 and SAMN20000888, respectively.
Additional information
Funding
References
- Andrews S. 2014. FastQC: q quality control tool for high throughput sequence data. http://wwwbioinformaticsbabrahamacuk/projects/fastqc/.
- Bankevich A, Nurk S, Antipov D, Gurevich A, Dvorkin M, Kulikov A, Lesin V, Nikolenko S, Pham S, Prjibelski A, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Bhagyasri Y, Ali P, Raja M, Reddy N, Praveen D, Mounika K, Latha D, Parameshwari N. 2019. Determination of in-vitro anti microbial activity and anti-diabetic activity of Ixora chinensis. AJPHR. 7(3):6–12.
- Chen L-Y, Chu C-Y, Huang M-C. 2003. Inflorescence and flower development in Chinese Ixora. JASHS. 128(1):23–28.
- Kim S-M, Kuzuyama T, Chang Y-J, Kim S-U. 2006. Cloning and characterization of 2-C-methyl-D:-erythritol 2,4-cyclodiphosphate synthase (MECS) gene from Ginkgo biloba. Plant Cell Rep. 25(8):829–835. doi:https://doi.org/10.1007/s00299-006-0136-3. 16528563
- Li T, Cai H, Wang T, Fu Y, Yang W, Zhao A, Cui Z, Wang J. 2019. Plant regeneration in Ixora chinensis from young leaves. Plant Cell Tiss Organ Cult. 139(3):605–608.
- Ly S, Garavito A, De Block P, Asselman P, Guyeux C, Charr J-C, Janssens S, Mouly A, Hamon P, Guyot R. 2020. Chloroplast genomes of Rubiaceae: comparative genomics and molecular phylogeny in subfamily Ixoroideae. PLoS One. 15(4):e0232295.
- Rastogi RM, Mehrotra BN. 1990. Compendium of Indian medicinal plants. Vol. 1. Lucknow: Central Drug Research Institute and Publications & Information Directorate, New Delhi; p. 388–389.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones E, Fischer A, Bock R, Greiner S. 2017. GeSeq - versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.