Abstract
The complete mitochondrial genome of Sillaginopsis panijus has been determined for the first time using Sanger Dideoxy DNA sequencing. The mitogenome is a circular molecule of 16,529 bp in length. It contains 37 mitochondrial genes (13 protein-coding genes, two ribosomal RNA, and 22 transfer RNA) and a control region as other bony fishes. In the phylogenetic analysis using 12H-strand protein-coding genes, monotypic S. panijus is situated separately from the genus Sillago. The present phylogeny supports its taxonomic position according to morphology and will be helpful for evolutionary analysis.
The Gangetic whiting, Sillaginopsis panijus (Hamilton-Buchanan Citation1822), is the only species of the genus Sillaginopsis. It is well documented as a commercial fish species due to its deliciousness, white flesh and few bones. It is distributed in the Northeastern Indian Ocean, including India, Bangladesh and Myanmar (Bay of Bengal), southward to Malaysia, and hardly to the Indonesian Archipelago (Andaman Sea) (McKay Citation1992). Its taxonomic description (Dutt and Sujatha Citation1980) and phylogeny (Kaga Citation2013) are mainly based on morphological characteristics. However, there were only partial COI and 12S rRNA sequences available in GenBank. Later an incomplete mitogenome (accession no. AP006802) (without a control region) was made available in GenBank before the release of data of this study. The present study sequenced and characterized its mitogenome and explored the phylogenetic relationship. The study will give supportive genetic information for future research in genetics and phylogenetics.
The specimen was collected from the Cox's Bazar, Bangladesh (21.452° N, 91.964° E) on 2nd October 2018. Muscle tissue (Voucher number, FEL_OUC142276) was deposited in the Fishery Ecology Laboratory, Fisheries College, Ocean University of China (http://eweb.ouc.edu.cn, Jia guang Xiao is the contact person [email protected]). Genomic DNA was extracted by proteinase K digestion followed by a standard phenol-chloroform method (Sambrook et al. Citation1989). A PCR-based primer walking method was used to ascertain the mitogenome. SeqMan (DNAStar, USA) software was used to manually correct and align the sequences. The start and end positions of each gene were determined by comparing with the complete mitochondrial DNA sequences of the Sillaginidae species published in GenBank. The tRNAscan-SE 2.0 (Lowe and Chan Citation2016) was used to identify tRNA genes. The location of the control area was determined through the position of tRNA-Phe and tRNA-Pro.
Maximum likelihood (ML) and Bayesian inference (BI) methods were carried out on concatenated sequences of protein-coding genes (PCGs) (except ND6) of eight Sillaginidae species to reveal the phylogenetic relationship. ClustalX 2.1 (Larkin et al. Citation2007) was used to align the sequences. By comparing the Akaike Information Criterion value in jModelTest v2.1.10 (Darriba et al. Citation2012), the GTR + I + G model was selected as the best to describe the substitution pattern. The PAUP* 4.0 (Swofford Citation2002) was used to perform ML analysis with 1000 bootstrap replications. Three partitioned (first, second and third codon positions of PCGs) BI analysis was done by MrBayes v. 3.2.6 (Ronquist et al. Citation2012).
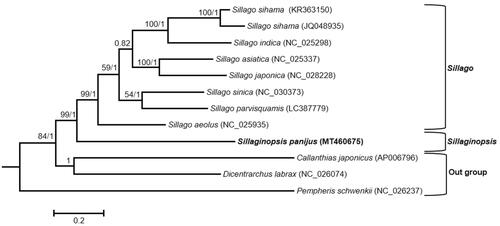
The mitogenome of S. panijus was sequenced to be 16,529 bp in length (GenBank accession no. MT460675). It consists of 13 typical vertebrate PCGs, 22 transfer RNA genes, two ribosomal RNA genes (12S rRNA and 16S rRNA), and two non-coding regions (control region and L-strand replication origin). It has shown the similarity with the canonical organization of the fish mitogenome in both gene content and order (Boore Citation1999). The encoding genes were located on the heavy strand with the exception of ND6 and eight tRNA (tRNA-Gln, tRNA-Ala, tRNA-Asn, tRNA-Cys, tRNA-Tyr, tRNA-Ser, tRNA-Glu and tRNA-Pro) genes were transcribed from the light strand. The overall nucleotide composition was estimated to be 27.97% T, 27.85% C, 26.18% A and 18.00% G, with anti-G bias and a slight excess of AT (54.15%). The 13 PCGs were a total of 11,443 bp in size, accounting for 69.23% of the whole mitogenome. All PCGs except COI use ATG as the start codon, while COI uses GTG as the start codon. Seven PCGs have the complete stop codon TAA (COI, ATPase8, COIII, ND4L and ND6) or TAG (ND1 and ND5). While the other six have the incomplete stop codon TA (ND2, ATPase6) or T (COII, ND3, ND4 and Cytb). In the phylogenetic tree, species of the genus Sillago clustered together. On the contrary, S. panijus was situated outside the clade of the genus Sillago that supported its morphological division at the genus level ().
Figure 1. Phylogenetic tree using BI and ML methods among eight Sillaginidae species based on concatenated sequences of the 12 PCGs. The Bayesian topology was similar to the result of the maximum likelihood tree. Bootstrap support values/Bayesian posterior probabilities were displayed at branch nodes. Three species of the suborder Percoidei, Callanthias japonicus, Dicentrarchus labrax and Pempheris schwenkii, were selected as outgroup species.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are publicly available in [GenBank accession number MT460675, https://www.ncbi.nlm.nih.gov/nuccore/MT460675].
Additional information
Funding
References
- Boore JL. 1999. Animal mitochondrial genomes. Nucleic Acids Res. 27(8):1767–1780.
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9(8):772.
- Dutt S, Sujatha K. 1980. On the seven species of the family Sillaginidae from Indian waters. Mahasagar – Bull Natl Inst Oceanogr. 13(4):371–375.
- Hamilton-Buchanan F. 1822. An account of the fishes found in the River Ganges and its branches. Edinburgh and London: Archibald Constable and Company; p. 405.
- Kaga T. 2013. Phylogenetic systematics of the family Sillaginidae (Percomorpha: order Perciformes). Zootaxa. 3642(1):1–105.
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics. 23(21):2947–2948.
- Lowe TM, Chan PP. 2016. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44(W1):W54–W57.
- McKay RJ. 1992. FAO Species Catalogue. Vol. 14. Sillaginid fishes of the world (family Sillaginidae). An annotated and illustrated catalogue of the Sillago, smelt or Indo-Pacific whiting species known to date. Rome: FAO. FAO Fish Synop. 125(14):87.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542.
- Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual. 2nd ed. New York: Cold Spring Harbor Laboratory Press.
- Swofford DL. 2002. PAUP* 4.0: phylogenetic analysis using parsimony. Sunderland (MA): Sinauer Associates Inc.
