Abstract
The complete chloroplast genome of Sparganium glomeratum was sequenced and assembled in this study. The circular genome is 160,391 bp in length and exhibits a typical quadripartite structure with a large single-copy (LSC, 87,660 bp) and small single-copy (SSC, 18,721 bp) regions, separated by a pair of inverted repeats (IRs, 27,005 bp). The cp genome contains 113 unique genes, including 79 protein-coding, 30 tRNA, and four rRNA genes. The phylogenetic analysis within the Poales showed that Sparganium is monophyletic and most closely related to Typha. Within Sparganium, S. glomeratum is sister to the clade of S. stoloniferum and S. euricarpum. The work reported here will provide useful information for the evolutionary studies on the genus of Sparganium.
Sparganium L. (Typhaceae) is an aquatic freshwater genus including approximately 14 species. Representatives of the genus occupy aquatic habitats primarily in temperate and cool regions, and several species show circumboreal distributions (Cook and Nicholls Citation1986, Citation1987). Species of Sparganium are ecologically important in aquatic communities, providing cover and food for a variety of waterfowl and mammals (Fassett Citation1940). Sparganium glomeratum (Laest. ex Beurl.) Beurl. is morphologically different from other species by its male and female heads, which are closely near each other on the inflorescence rachis (Sun and Simpson Citation2010). S. glomeratum is polymorphic, occurring either as a floating or emergent plant. To date, the complete chloroplast genomes of two emergent Sparganium taxa have been sequenced, S. erectum subsp. stoloniferum and S. eurycarpum subsp. coreanum (Gil et al. Citation2019; Su et al. Citation2019). As Sparganium is of biogeographical, phylogenetic, and evolutionary interest, more genetic resources are necessary for further understand the systematics and phylogenetic history of the genus. Here, we report the complete cp genome of S. glomeratum and reconstruct a phylogenetic tree to confirm its position and the monophyly of Sparganium.
Fresh leaves of S. glomeratum were collected from Fusong county (127°33.174′E, 42°1.146′N) in Jilin Province of China, and dried with silica gel. The specimens were deposited in the herbarium of Wuhan University (www.whu.edu.cn, Xinwei Xu, [email protected]) under the voucher number Xu3795. Total genomic DNA was extracted from ∼3 mg of tissue using the DNA Plantzol Reagent (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s protocol. Purified DNA was sheared into ∼500 bp fragments, and the paired-end sequencing libraries were constructed according to the Illumina standard protocol (Illumina, San Diego, CA, USA). The genomic DNA of the sample was sequenced using an Illumina BGISEQ-500 (Illumina, San Diego, CA, USA) at the Beijing Genomics Institute (BGI; Shenzhen, China). After removing adapters, Illumina paired-end sequencing generated a total of 20,631,044 clean reads. These clean reads were then used to assemble the cp genomes using NOVOPlasty4.2 (Dierckxsens et al. Citation2017), with a subunit of the photosystem II (psbA) gene from S. stoloniferum (GenBank accession no. NC_044634) as the seed. The chloroplast genome annotation was performed with Geneious Prime v2020.0.5 (Kearse et al. Citation2012), with the cp genome of S. stoloniferum (GenBank accession no. NC_044634) as the reference. Where necessary, the start and stop codon positions and the boundaries between introns and exons were manually corrected. The annotated complete cp genomes of S. glomeratum was deposited in GenBank under accession no. MZ681495.
The chloroplast genome of S. glomeratum is a circular DNA molecule of 160,391 bp in length. The cp genome has a typical quadripartite structure, consisting of a pair of inverted repeats (IRa and IRb, 27,005 bp) separated by a large single-copy region (LSC, 87,660 bp) and a small single-copy region (SSC, 18,721 bp). The overall GC content was 36.9%. The IR regions had a higher GC content (42.4%) than the LSC (34.9%) and SSC (30.9%) regions. The cp genome encoded a set of 113 unique genes, including 79 protein-coding, 30 tRNA genes, and 4 rRNA genes. Twenty genes, including eight protein-coding genes, eight tRNA genes, and four rRNA genes, were duplicated in the IR region. Among the genes identified, nine genes (atpF, ndhA, ndhB, petB, petD, rpl2, rpl16, rpoC1, and rps16) contained one intron and three genes (clpP, rps12, and ycf3) comprised of two introns. In addition, 20 genes were duplicated in the IR regions, and ycf1 its pseudogene.
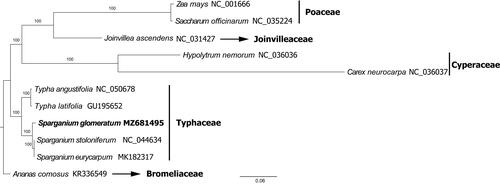
The cp genomes of eleven species of Poales were downloaded from GenBank for the phylogenetic analysis. The sequences were aligned using the auto setting in MAFFT v7.3 (Katoh and Standley Citation2013). The phylogenetic tree was constructed using IQTREE v1.6.7 (Nguyen et al. Citation2015), with the best selected GTR + F + R2 model and 1000 fast bootstrap replicates. Ananas comosus (L.) Merr. was designated as the outgroup. The result confirmed the sister relationship of genus Typha and Sparganium, supporting their inclusion in the Typhaceae (). Within Sparganium, S. glomeratum was fully resolved in a sister position to the two erect species that produce distigmatic flowers and bilocular fruits, these are S. stoloniferum and S. euricarpum. These phylogenetic findings are in accordance with the results of Sulman et al. (Citation2013) based on chloroplast and nuclear DNA marker analysis.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession no. MZ681495. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA750266, SRR15312016, and SAMN20447564, respectively.
Additional information
Funding
References
- Cook CDK, Nicholls MS. 1986. A monographic study of the genus Sparganium (Sparganiaceae). Part 1, Subgenus Xanthosparganium Holmberg. Bot Helv. 96:213–267.
- Cook CDK, Nicholls MS. 1987. A monographic study of the genus Sparganium (Sparganiaceae). Part 2, Subgenus Sparganium. Bot Helv. 97:1–44.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Fassett NC. 1940. A manual of aquatic plants. Madison (USA): University of Wisconsin Press.
- Gil HY, Ha YH, Choi KS, Lee JS, Chang KS, Choi K. 2019. The chloroplast genome sequence of an aquatic plant, Sparganium eurycarpum subsp. coreanum (Typhaceae). Mitochondrial DNA Part B. 4(1):684–685.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Sulman JD, Drew BT, Drummond C, Hayasaka E, Sytsma KJ. 2013. Systematics, biogeography, and character evolution of Sparganium (Typhaceae): diversification of a widespread, aquatic lineage. Am J Bot. 100(10):2023–2039.
- Sun K, Simpson DA. 2010. Typhaceae. In: Wu ZY, Raven PH, Hong DY, editors. Flora of China. Vol. 23. Acoraceae through Cyperaceae. Beijing (China): Science Press; St, Louis (MI): Missouri Botanical Garden Press; p. 158–163.
- Su T, Yang JX, Lin YG, Kang N, Hu GX. 2019. Characterization of the complete chloroplast genome of Sparganium stoloniferum (Poales: Typhaceae) and phylogenetic analysis. Mitochondrial DNA Part B. 4(1):1402–1403.