Abstract
The complete mitochondrial DNA sequence of Apogonichthyoides taeniatus (Cuvier, 1828) is determined. The mitochondrial genome is 17,050 in length and has the same composition and gene order like most other vertebrates. The phylogenetic analysis based on 13 concatenated PCGs nucleotide sequences among 20 species showed that this species has high support with the sister branch Jaydia lineata. Our findings provide useful information for phylogenetic and evolutionary research of Kurtiformes species.
The two-belt cardinal and striped cardinalfish Apogonichthyoides taeniatus (Cuvier, 1828), belong to Apogoninae, Kurtiformes. It distributes generally in the Indian Ocean-western and South China Sea (Masuda et al. Citation1984). A. taeniatus generally inhabit coastal coral reefs and mangrove waters. The biology of A. taeniatus is poorly studied, with only morphological and behavioral studies reported (Thresher et al. Citation1984; Tortonese Citation1986). In the present study, we characterize the complete mitochondrial genome DNA (mtDNA) sequence of A. taeniatus.
A. taeniatus were collected from Hailing Island, Yangjiang city, Guangdong province, China (111°52′00″E, 21°40′00″N). The genomic DNA extracts from the standard phenol/chloroform method (Sambrook and Russel 2002). The method for preparing and sequencing the DNA library is referred to Zhu et al. (Citation2017a, Citation2017b). Twenty-five primer pairs designed from the mitogenome of related species were used to amplify the A. taeniatus mitogenome (Supplementary Table 1). The obtained partial sequences of A. taeniatus were assembled using Geneious R11.1.5 (Kearse et al. Citation2012), and the final complete mitogenome was annotated based on the Apogon semilineatus mitogenome (GenBank: AP005996). The specimens and DNA were deposited at the College of Marine Sciences, South China Agricultural University (https://www.scau.edu.cn, Min Yang, [email protected]) under voucher number SCAU-AT-0404.
The complete mtDNA is 17, 050 bp in length (Genbank Accession number: MN699562), with a high A + T content (51.80%). A (27.85%), T (30.97%), C (19.18%), and G (21.99%) are in D-loop region. There are eight overlapping regions, ranging from 1 to 10 bp. And 13 intergenic regions exist in this genome, ranging from 1 to 35 bp. The gene content is equal to many other teleost mtDNA (Yagishita et al. Citation2009; Zhang et al. Citation2016; Zhu et al. Citation2017a), comprising 13 protein-coding genes (PCGs), 22 tRNAs, 2 rRNAs, and a control region.
Only one PCGs (ND6), and eight tRNA genes (Gln, Ala, Asn, Cys, Tyr, Ser, Glu, and Pro) are encoded by L strand, while two rRNA genes (12S rRNA and 16S rRNA), twelve PCGs (ND1, ND2, COX1, COXII, ATP8, ATP6, COXIII, ND3, ND4L, ND5, and Cytb), fourteen tRNA genes (Phe, Val, Leu, Ile, Met, Trp, Asp, Lys, Gly, Arg, His, Ser, Leu, and Thr), and a non-coding region (D-loop region) are encoded by H strand. Twelve of thirteen PCGs start with the representative initiation codon ATG, only COX1 starts with GTG. Nine PCGs initial TAA and ND2 use AGA as a stop codon, ND6 use AGG as a stop codon, whereas COXII, ND4, and Cytb use incomplete stop codon, T-. It is analogously with that in Toxotes chatareus, Elagatis bipinnulata, and Lutjanus carponotatus (Yagishita et al. Citation2009; Ma et al. Citation2017; Kim et al. Citation2019).
There are 22 tRNAs genes, ranging from 66 bp (tRNACys) to 75 bp (tRNALeu). The non-coding region is A + T enrichment region, which is related to transcription and replication (Clayton Citation1992). The non-coding region is located between tRNAPro and tRNAPhe genes, is 1314 bp length with high A + T content (58.82%). All the tRNAs can be folded into the representative cloverleaf secondary structures.
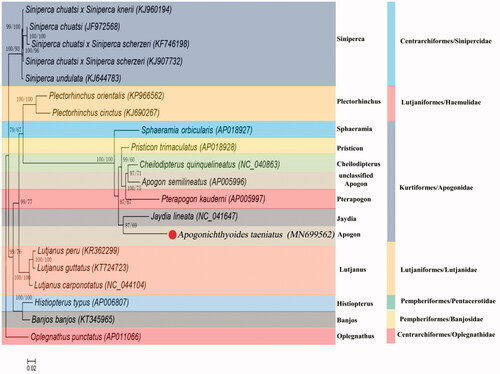
To investigate phylogenetic relationships between Centrarchiformes, Lutjaniformes, Pempheriformes, and Kurtiformes, a phylogenetic tree is constructed by MEGA 7.0 based on 13 tandem PCGs nucleotide sequences with Maximum likelihood (ML) method and MtMam + I + G + F model (Kumar et al. Citation2016). Because both two species belong to Kurtiformes, A. taeniatus is grouped with J. lineata as the sister species (). The genetic distance among Kurtiformes/Apogonidae species was also calculated basing on the complete mtDNA sequence by MEGA 7.0, and the results showed that the species in descending order of genetic distance from A. taeniatus were P. trimaculatus, J. lineata, C. quinquelineatus, A. semilineatus, S. orbicularis, P. kaudemi (Supplementary Table 2). The A. taeniatus mtDNA will provide additional genetic information for genetic analyses in future study.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in the Genbank of NCBI at https://www.ncbi.nlm.nih.gov/, reference number MN699562.
Additional information
Funding
References
- Clayton DA. 1992. Transcription and replication of animal mitochondrial DNAs. Int Rev Cytol. 141:217–232.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Kim G, Lee JH, Alam MJ, Lee SR, Andriyono S. 2019. Complete mitochondrial genome of Spanish flag snapper, Lutjanus carponotatus (Perciformes: Lutjanidae). Mitochondrial DNA B Resour. 4(1):568–569.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Ma C, Ma H, Zhang H, Feng C, Wei H, Wang W, Chen W, Zhang F, Ma L. 2017. The complete mitochondrial genome sequence and gene organization of the rainbow runner (Elagatis bipinnulata) (Perciformes: Carangidae). Mitochondrial DNA A DNA Mapp Seq Anal. 28(1):5–6.
- Masuda H, Amaoka K, Araga C, Uyeno T, Yoshino T. 1984. The fishes of the Japanese archipelago. Vol. 1. Tokyo (Japan): Tokai University Press; p. 437.
- Sambrook JD, Russell W. 2000. Molecular cloning: a laboratory manual. 2nd edn. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press.
- Thresher RE. 1984. Reproduction in reef fishes. Neptune City, New Jersey: T.F.H. Publications, Inc. Ltd.; p. 399.
- Tortonese E. 1986. Fishes of the north-eastern Atlantic and the Mediterranean. Vol. 2. Paris: UNESCO; p. 803–809.
- Yagishita N, Miya M, Yamanoue Y, Shirai SM, Nakayama K, Suzuki N, Satoh TP, Mabuchi K, Nishida M, Nakabo T. 2009. Mitogenomic evaluation of the unique facial nerve pattern as a phylogenetic marker within the percifom fishes (Teleostei: Percomorpha). Mol Phylogenet Evol. 53(1):258–266.
- Zhang DC, Wang L, Guo H, Ma Z, Zhang N, Lin J, Jiang SG. 2016. Complete mitochondrial genome of Florida pompano Trachinotus carolinus (Teleostei, Carangidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27(1):597–598.
- Zhu KC, Wu N, Sun XX, Guo HY, Zhang N, Jiang SG, Zhang DC. 2017a. Characterization of complete mitochondrial genome of fives tripe wrasse (Thalassoma quinquevittatum, Lay & Bennett, 1839) and phylogenetic analysis. Gene. 598:71–78.
- Zhu KC, Liang YY, Wu N, Guo HY, Zhang N, Jiang SG, Zhang DC. 2017b. Sequencing and characterization of the complete mitochondrial genome of Japanese Swellshark (Cephalloscyllium umbratile). Sci Rep. 7(1):15299.